SRGAP2 and Its Human-Specific Paralog Co-Regulate the Development of Excitatory and Inhibitory Synapses
- PMID: 27373832
- PMCID: PMC5385270
- DOI: 10.1016/j.neuron.2016.06.013
SRGAP2 and Its Human-Specific Paralog Co-Regulate the Development of Excitatory and Inhibitory Synapses
Abstract
The proper function of neural circuits requires spatially and temporally balanced development of excitatory and inhibitory synapses. However, the molecular mechanisms coordinating excitatory and inhibitory synaptogenesis remain unknown. Here we demonstrate that SRGAP2A and its human-specific paralog SRGAP2C co-regulate the development of excitatory and inhibitory synapses in cortical pyramidal neurons in vivo. SRGAP2A promotes synaptic maturation, and ultimately the synaptic accumulation of AMPA and GABAA receptors, by interacting with key components of both excitatory and inhibitory postsynaptic scaffolds, Homer and Gephyrin. Furthermore, SRGAP2A limits the density of both types of synapses via its Rac1-GAP activity. SRGAP2C inhibits all identified functions of SRGAP2A, protracting the maturation and increasing the density of excitatory and inhibitory synapses. Our results uncover a molecular mechanism coordinating critical features of synaptic development and suggest that human-specific duplication of SRGAP2 might have contributed to the emergence of unique traits of human neurons while preserving the excitation/inhibition balance.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





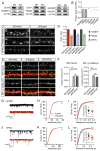
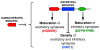
Comment in
-
Filling the (SR)GAP in Excitatory/Inhibitory Balance.Neuron. 2016 Jul 20;91(2):205-7. doi: 10.1016/j.neuron.2016.07.008. Neuron. 2016. PMID: 27477010
References
-
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 2007;30:79–97. - PubMed
-
- Benavides-Piccione R, Ballesteros-Yanez I, DeFelipe J, Yuste R. Cortical area and species differences in dendritic spine morphology. J Neurocytol. 2002;31:337–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

