HPIDB 2.0: a curated database for host-pathogen interactions
- PMID: 27374121
- PMCID: PMC4930832
- DOI: 10.1093/database/baw103
HPIDB 2.0: a curated database for host-pathogen interactions
Abstract
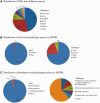
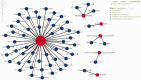
Identification and analysis of host-pathogen interactions (HPI) is essential to study infectious diseases. However, HPI data are sparse in existing molecular interaction databases, especially for agricultural host-pathogen systems. Therefore, resources that annotate, predict and display the HPI that underpin infectious diseases are critical for developing novel intervention strategies. HPIDB 2.0 (http://www.agbase.msstate.edu/hpi/main.html) is a resource for HPI data, and contains 45, 238 manually curated entries in the current release. Since the first description of the database in 2010, multiple enhancements to HPIDB data and interface services were made that are described here. Notably, HPIDB 2.0 now provides targeted biocuration of molecular interaction data. As a member of the International Molecular Exchange consortium, annotations provided by HPIDB 2.0 curators meet community standards to provide detailed contextual experimental information and facilitate data sharing. Moreover, HPIDB 2.0 provides access to rapidly available community annotations that capture minimum molecular interaction information to address immediate researcher needs for HPI network analysis. In addition to curation, HPIDB 2.0 integrates HPI from existing external sources and contains tools to infer additional HPI where annotated data are scarce. Compared to other interaction databases, our data collection approach ensures HPIDB 2.0 users access the most comprehensive HPI data from a wide range of pathogens and their hosts (594 pathogen and 70 host species, as of February 2016). Improvements also include enhanced search capacity, addition of Gene Ontology functional information, and implementation of network visualization. The changes made to HPIDB 2.0 content and interface ensure that users, especially agricultural researchers, are able to easily access and analyse high quality, comprehensive HPI data. All HPIDB 2.0 data are updated regularly, are publically available for direct download, and are disseminated to other molecular interaction resources.Database URL: http://www.agbase.msstate.edu/hpi/main.html.
© The Author(s) 2016. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

