Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan
- PMID: 27374331
- PMCID: PMC5534359
- DOI: 10.1016/j.cell.2016.05.076
Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan
Abstract
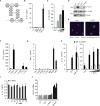
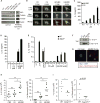
Degradation of Gram-positive bacterial cell wall peptidoglycan in macrophage and dendritic cell phagosomes leads to activation of the NLRP3 inflammasome, a cytosolic complex that regulates processing and secretion of interleukin (IL)-1β and IL-18. While many inflammatory responses to peptidoglycan are mediated by detection of its muramyl dipeptide component in the cytosol by NOD2, we report here that NLRP3 inflammasome activation is caused by release of N-acetylglucosamine that is detected in the cytosol by the glycolytic enzyme hexokinase. Inhibition of hexokinase by N-acetylglucosamine causes its dissociation from mitochondria outer membranes, and we found that this is sufficient to activate the NLRP3 inflammasome. In addition, we observed that glycolytic inhibitors and metabolic conditions affecting hexokinase function and localization induce inflammasome activation. While previous studies have demonstrated that signaling by pattern recognition receptors can regulate metabolic processes, this study shows that a metabolic enzyme can act as a pattern recognition receptor. PAPERCLIP.
Copyright © 2016. Published by Elsevier Inc.
Figures
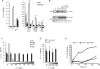



Comment in
-
Immunometabolism: Innate sensing role for metabolic enzyme.Nat Rev Immunol. 2016 Aug;16(8):463. doi: 10.1038/nri.2016.84. Epub 2016 Jul 18. Nat Rev Immunol. 2016. PMID: 27424775 No abstract available.
-
When Hexokinase Gets that NAG-ing Feeling….Cell Metab. 2016 Aug 9;24(2):198-200. doi: 10.1016/j.cmet.2016.07.021. Cell Metab. 2016. PMID: 27508867 Free PMC article.
-
NAGging Hexokinase PEPs up NLRP3.Cell Host Microbe. 2016 Aug 10;20(2):130-2. doi: 10.1016/j.chom.2016.07.017. Cell Host Microbe. 2016. PMID: 27512900
References
-
- Aleshin AE, Zeng C, Bartunik HD, Fromm HJ, Honzatko RB. Regulation of hexokinase I: crystal structure of recombinant human brain hexokinase complexed with glucose and phosphate. J Mol Biol. 1998a;282:345–357. - PubMed
-
- Aleshin AE, Zeng C, Bourenkov GP, Bartunik HD, Fromm HJ, Honzatko RB. The mechanism of regulation of hexokinase: new insights from the crystal structure of recombinant human brain hexokinase complexed with glucose and glucose-6-phosphate. Structure. 1998b;6:39–50. - PubMed
-
- Berg JM, Tymoczko JL, Stryer L, editors. Biochemistry. Fifth. New York: W. H. Freeman; 2002. The glycolytic pathway is tightly controlled. Section 16.2. http://www.ncbi.nlm.nih.gov/books/NBK21154/
-
- Chiara F, Castellaro D, Marin O, Petronilli V, Brusilow WS, Juhaszova M, Sollott SJ, Forte M, Bernardi P, Rasola A. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLoS ONE. 2008;3:e1852. - PMC - PubMed
-
- da-Silva WS, Gómez-Puyou A, de Gómez-Puyou MT, Moreno-Sanchez R, De Felice FG, de Meis L, Oliveira MF, Galina A. Mitochondrial bound hexokinase activity as a preventive antioxidant defense: steady-state ADP formation as a regulatory mechanism of membrane potential and reactive oxygen species generation in mitochondria. J Biol Chem. 2004;279:39846–39855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

