Fungal and Prokaryotic Activities in the Marine Subsurface Biosphere at Peru Margin and Canterbury Basin Inferred from RNA-Based Analyses and Microscopy
- PMID: 27375571
- PMCID: PMC4899926
- DOI: 10.3389/fmicb.2016.00846
Fungal and Prokaryotic Activities in the Marine Subsurface Biosphere at Peru Margin and Canterbury Basin Inferred from RNA-Based Analyses and Microscopy
Abstract
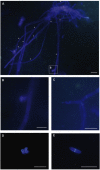
The deep sedimentary biosphere, extending 100s of meters below the seafloor harbors unexpected diversity of Bacteria, Archaea, and microbial eukaryotes. Far less is known about microbial eukaryotes in subsurface habitats, albeit several studies have indicated that fungi dominate microbial eukaryotic communities and fungal molecular signatures (of both yeasts and filamentous forms) have been detected in samples as deep as 1740 mbsf. Here, we compare and contrast fungal ribosomal RNA gene signatures and whole community metatranscriptomes present in sediment core samples from 6 and 95 mbsf from Peru Margin site 1229A and from samples from 12 and 345 mbsf from Canterbury Basin site U1352. The metatranscriptome analyses reveal higher relative expression of amino acid and peptide transporters in the less nutrient rich Canterbury Basin sediments compared to the nutrient rich Peru Margin, and higher expression of motility genes in the Peru Margin samples. Higher expression of genes associated with metals transporters and antibiotic resistance and production was detected in Canterbury Basin sediments. A poly-A focused metatranscriptome produced for the Canterbury Basin sample from 345 mbsf provides further evidence for active fungal communities in the subsurface in the form of fungal-associated transcripts for metabolic and cellular processes, cell and membrane functions, and catalytic activities. Fungal communities at comparable depths at the two geographically separated locations appear dominated by distinct taxa. Differences in taxonomic composition and expression of genes associated with particular metabolic activities may be a function of sediment organic content as well as oceanic province. Microscopic analysis of Canterbury Basin sediment samples from 4 and 403 mbsf produced visualizations of septate fungal filaments, branching fungi, conidiogenesis, and spores. These images provide another important line of evidence supporting the occurrence and activity of fungi in the deep subseafloor biosphere.
Keywords: DNA; calcofluor white; fungi; iTAG; mRNA; metatranscriptome; rRNA operon; sediment.
Figures




References
-
- Adeyemi A. O. (2009). Bioaccumulation of arsenic in fungi. Am. J. Environ. Sci. 5 364–370. 10.3844/ajessp.2009.364.370 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

