Klebsiella pneumoniae Asparagine tDNAs Are Integration Hotspots for Different Genomic Islands Encoding Microcin E492 Production Determinants and Other Putative Virulence Factors Present in Hypervirulent Strains
- PMID: 27375573
- PMCID: PMC4891358
- DOI: 10.3389/fmicb.2016.00849
Klebsiella pneumoniae Asparagine tDNAs Are Integration Hotspots for Different Genomic Islands Encoding Microcin E492 Production Determinants and Other Putative Virulence Factors Present in Hypervirulent Strains
Abstract
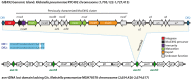
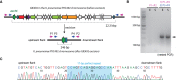
Due to the developing of multi-resistant and invasive hypervirulent strains, Klebsiella pneumoniae has become one of the most urgent bacterial pathogen threats in the last years. Genomic comparison of a growing number of sequenced isolates has allowed the identification of putative virulence factors, proposed to be acquirable mainly through horizontal gene transfer. In particular, those related with synthesizing the antibacterial peptide microcin E492 (MccE492) and salmochelin siderophores were found to be highly prevalent among hypervirulent strains. The determinants for the production of both molecules were first reported as part of a 13-kbp segment of K. pneumoniae RYC492 chromosome, and were cloned and characterized in E. coli. However, the genomic context of this segment in K. pneumoniae remained uncharacterized. In this work, we provided experimental and bioinformatics evidence indicating that the MccE492 cluster is part of a highly conserved 23-kbp genomic island (GI) named GIE492, that was integrated in a specific asparagine-tRNA gene (asn-tDNA) and was found in a high proportion of isolates from liver abscesses sampled around the world. This element resulted to be unstable and its excision frequency increased after treating bacteria with mitomycin C and upon the overexpression of the island-encoded integrase. Besides the MccE492 genetic cluster, it invariably included an integrase-coding gene, at least seven protein-coding genes of unknown function, and a putative transfer origin that possibly allows this GI to be mobilized through conjugation. In addition, we analyzed the asn-tDNA loci of all the available K. pneumoniae assembled chromosomes to evaluate them as GI-integration sites. Remarkably, 73% of the strains harbored at least one GI integrated in one of the four asn-tDNA present in this species, confirming them as integration hotspots. Each of these tDNAs was occupied with different frequencies, although they were 100% identical. Also, we identified a total of 47 asn-tDNA-associated GIs that were classified into 12 groups of homology differing in theencoded functionalities but sharing with GIE492 a conserved recombination module and potentially its mobility features. Most of these GIs encoded factors with proven or potential role in pathogenesis, constituting a major reservoir of virulence factors in this species.
Keywords: asparagine tRNA gene; hypervirulent Klebsiella pneumoniae; liver abscess; microcin E492; pathogenicity island; salmochelin.
Figures

References
-
- Ahmed S., Awosika J., Baldwin C., Bishop-Lilly K. A., Biswas B., Broomall S., et al. (2012). Genomic comparison of Escherichia coli O104:H4 Isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS ONE 7:e48228 10.1371/journal.pone.0048228 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

