Detecting the Hidden Properties of Immunological Data and Predicting the Mortality Risks of Infectious Syndromes
- PMID: 27375617
- PMCID: PMC4901050
- DOI: 10.3389/fimmu.2016.00217
Detecting the Hidden Properties of Immunological Data and Predicting the Mortality Risks of Infectious Syndromes
Abstract
Background: To extract more information, the properties of infectious disease data, including hidden relationships, could be considered. Here, blood leukocyte data were explored to elucidate whether hidden information, if uncovered, could forecast mortality.
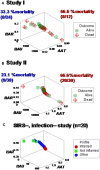
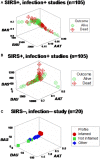
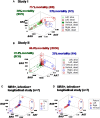
Methods: Three sets of individuals (n = 132) were investigated, from whom blood leukocyte profiles and microbial tests were conducted (i) cross-sectional analyses performed at admission (before bacteriological tests were completed) from two groups of hospital patients, randomly selected at different time periods, who met septic criteria [confirmed infection and at least three systemic inflammatory response syndrome (SIRS) criteria] but lacked chronic conditions (study I, n = 36; and study II, n = 69); (ii) a similar group, tested over 3 days (n = 7); and (iii) non-infected, SIRS-negative individuals, tested once (n = 20). The data were analyzed by (i) a method that creates complex data combinations, which, based on graphic patterns, partitions the data into subsets and (ii) an approach that does not partition the data. Admission data from SIRS+/infection+ patients were related to 30-day, in-hospital mortality.
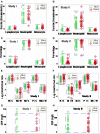
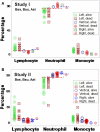
Results: The non-partitioning approach was not informative: in both study I and study II, the leukocyte data intervals of non-survivors and survivors overlapped. In contrast, the combinatorial method distinguished two subsets that, later, showed twofold (or larger) differences in mortality. While the two subsets did not differ in gender, age, microbial species, or antimicrobial resistance, they revealed different immune profiles. Non-infected, SIRS-negative individuals did not express the high-mortality profile. Longitudinal data from septic patients displayed the pattern associated with the highest mortality within the first 24 h post-admission. Suggesting inflammation coexisted with immunosuppression, one high-mortality sub-subset displayed high neutrophil/lymphocyte ratio values and low lymphocyte percents. A second high-mortality subset showed monocyte-mediated deficiencies. Numerous within- and between-subset comparisons revealed statistically significantly different immune profiles.
Conclusion: While the analysis of non-partitioned data can result in information loss, complex (combinatorial) data structures can uncover hidden patterns, which guide data partitioning into subsets that differ in mortality rates and immune profiles. Such information can facilitate diagnostics, monitoring of disease dynamics, and evaluation of subset-specific, patient-specific therapies.
Keywords: complexity; immunomicrobial interactions; immunosuppression; pattern recognition; sepsis; visual.
Figures







References
-
- Berens P. CircStat: a MATLAB toolbox for circular statistics. J Stat Softw (2009) 31:1–21. 10.18637/jss.v031.i10 - DOI
-
- Gill J, Hangartner D. Circular data in political science and how to handle it. Polit Anal (2010) 18:316–36. 10.1093/pan/mpq009 - DOI
-
- Fisher R. Dispersion on a sphere. Proc R Soc Lond A (1953) 217:295–305. 10.1098/rspa.1953.0064 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

