Characterization and Expression Analysis of PtAGL24, a SHORT VEGETATIVE PHASE/AGAMOUS-LIKE 24 (SVP/AGL24)-Type MADS-Box Gene from Trifoliate Orange (Poncirus trifoliata L. Raf.)
- PMID: 27375669
- PMCID: PMC4901042
- DOI: 10.3389/fpls.2016.00823
Characterization and Expression Analysis of PtAGL24, a SHORT VEGETATIVE PHASE/AGAMOUS-LIKE 24 (SVP/AGL24)-Type MADS-Box Gene from Trifoliate Orange (Poncirus trifoliata L. Raf.)
Abstract
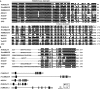
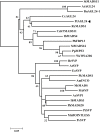
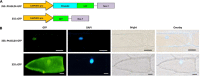
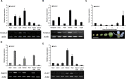
The transition from vegetative to reproductive growth in perennial woody plants does not occur until after several years of repeated seasonal changes and alternative growth. To better understand the molecular basis of flowering regulation in citrus, a MADS-box gene was isolated from trifoliate orange (precocious trifoliate orange, Poncirus trifoliata L. Raf.). Sequence alignment and phylogenetic analysis showed that the MADS-box gene is more closely related to the homologs of the AGAMOUS-LIKE 24 (AGL24) lineage than to any of the other MADS-box lineages known from Arabidopsis; it is named PtAGL24. Expression analysis indicated that PtAGL24 was widely expressed in the most organs of trifoliate orange, with the higher expression in mature flowers discovered by real-time PCR. Ectopic expression of PtAGL24 in wild-type Arabidopsis promoted early flowering and caused morphological changes in class I transgenic Arabidopsis. Yeast two-hybrid assay revealed that PtAGL24 interacted with Arabidopsis AtAGL24 and other partners of AtAGL24, suggesting that the abnormal morphology of PtAGL24 overexpression in transgenic Arabidopsis was likely due to the inappropriate interactions between exogenous and endogenous proteins. Also, PtAGL24 interacted with SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (PtSOC1) and APETALA1 (PtAP1) of citrus. These results suggest that PtAGL24 may play an important role in the process of floral transition but may have diverse functions in citrus development.
Keywords: MADS-box; PtAGL24; floral development; flowering; trifoliate orange.
Figures

Similar articles
-
PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants.Plant Mol Biol. 2010 Sep;74(1-2):129-42. doi: 10.1007/s11103-010-9660-1. Epub 2010 Jul 3. Plant Mol Biol. 2010. PMID: 20602150
-
PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuation in expression level.Planta. 2009 Mar;229(4):847-59. doi: 10.1007/s00425-008-0885-z. Epub 2009 Jan 6. Planta. 2009. PMID: 19125288
-
Molecular cloning and functional characterization of genes associated with flowering in citrus using an early-flowering trifoliate orange (Poncirus trifoliata L. Raf.) mutant.Plant Mol Biol. 2011 May;76(1-2):187-204. doi: 10.1007/s11103-011-9780-2. Epub 2011 May 1. Plant Mol Biol. 2011. PMID: 21533840
-
Evolution and expression of the MADS-box flowering transition genes AGAMOUS-like 24/SHORT VEGETATIVE PHASE with emphasis in selected Neotropical orchids.Cells Dev. 2021 Dec;168:203755. doi: 10.1016/j.cdev.2021.203755. Epub 2021 Nov 8. Cells Dev. 2021. PMID: 34758403
-
Identification of flowering-related genes between early flowering trifoliate orange mutant and wild-type trifoliate orange (Poncirus trifoliata L. Raf.) by suppression subtraction hybridization (SSH) and macroarray.Gene. 2009 Feb 1;430(1-2):95-104. doi: 10.1016/j.gene.2008.09.023. Epub 2008 Oct 1. Gene. 2009. PMID: 18930791
Cited by
-
WRKY transcription factor MdWRKY71 regulates flowering time in apple.Plant Mol Biol. 2025 Feb 13;115(2):32. doi: 10.1007/s11103-024-01544-8. Plant Mol Biol. 2025. PMID: 39945922
-
Characterization of Phytohormones and Transcriptomic Profiling of the Female and Male Inflorescence Development in Manchurian Walnut (Juglans mandshurica Maxim.).Int J Mol Sci. 2022 May 13;23(10):5433. doi: 10.3390/ijms23105433. Int J Mol Sci. 2022. PMID: 35628244 Free PMC article.
-
Isolation and Functional Characterization of the MADS-Box Gene AGAMOUS-LIKE 24 in Rubber Dandelion (Taraxacum kok-saghyz Rodin).Int J Mol Sci. 2025 Mar 4;26(5):2271. doi: 10.3390/ijms26052271. Int J Mol Sci. 2025. PMID: 40076890 Free PMC article.
-
IiSVP of Isatis indigotica can reduce the size and repress the development of floral organs.Plant Cell Rep. 2023 Mar;42(3):561-574. doi: 10.1007/s00299-023-02977-z. Epub 2023 Jan 7. Plant Cell Rep. 2023. PMID: 36609767
-
Association of extracellular dNTP utilization with a GmPAP1-like protein identified in cell wall proteomic analysis of soybean roots.J Exp Bot. 2018 Jan 23;69(3):603-617. doi: 10.1093/jxb/erx441. J Exp Bot. 2018. PMID: 29329437 Free PMC article.
References
-
- Bowman J. L., Alvarez J., Weigel D., Meyerowitz E. M., Smyth D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

