Activin A programs the differentiation of human TFH cells
- PMID: 27376469
- PMCID: PMC4955732
- DOI: 10.1038/ni.3494
Activin A programs the differentiation of human TFH cells
Erratum in
-
Erratum: Activin A programs the differentiation of human TFH cells.Nat Immunol. 2016 Sep 20;17(10):1235. doi: 10.1038/ni1016-1235d. Nat Immunol. 2016. PMID: 27648551 No abstract available.
Abstract
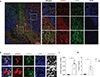
Follicular helper T cells (TFH cells) are CD4(+) T cells specialized in helping B cells and are associated both with protective antibody responses and autoimmune diseases. The promise of targeting TFH cells therapeutically has been limited by fragmentary understanding of extrinsic signals that regulate the differentiation of human TFH cells. A screen of a human protein library identified activin A as a potent regulator of TFH cell differentiation. Activin A orchestrated the expression of multiple genes associated with the TFH program, independently or in concert with additional signals. TFH cell programming by activin A was antagonized by the cytokine IL-2. Activin A's ability to drive TFH cell differentiation in vitro was conserved in non-human primates but not in mice. Finally, activin-A-induced TFH programming was dependent on signaling via SMAD2 and SMAD3 and was blocked by pharmacological inhibitors.
Figures




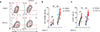

References
-
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

