Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi
- PMID: 27376574
- PMCID: PMC5084084
- DOI: 10.1038/nm.4139
Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi
Erratum in
-
Corrigendum: Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi.Nat Med. 2016 Oct 6;22(10):1192. doi: 10.1038/nm1016-1192b. Nat Med. 2016. PMID: 27711068 No abstract available.
Abstract
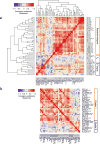
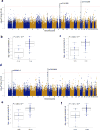
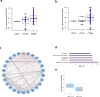
Little is known about the inter-individual variation of cytokine responses to different pathogens in healthy individuals. To systematically describe cytokine responses elicited by distinct pathogens and to determine the effect of genetic variation on cytokine production, we profiled cytokines produced by peripheral blood mononuclear cells from 197 individuals of European origin from the 200 Functional Genomics (200FG) cohort in the Human Functional Genomics Project (http://www.humanfunctionalgenomics.org), obtained over three different years. We compared bacteria- and fungi-induced cytokine profiles and found that most cytokine responses were organized around a physiological response to specific pathogens, rather than around a particular immune pathway or cytokine. We then correlated genome-wide single-nucleotide polymorphism (SNP) genotypes with cytokine abundance and identified six cytokine quantitative trait loci (QTLs). Among them, a cytokine QTL at the NAA35-GOLM1 locus markedly modulated interleukin (IL)-6 production in response to multiple pathogens and was associated with susceptibility to candidemia. Furthermore, the cytokine QTLs that we identified were enriched among SNPs previously associated with infectious diseases and heart diseases. These data reveal and begin to explain the variability in cytokine production by human immune cells in response to pathogens.
Figures



References
-
- Netea MG, Wijmenga C, O’Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nature immunology. 2012;13:535–542. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

