Nanoscale imaging of RNA with expansion microscopy
- PMID: 27376770
- PMCID: PMC4965288
- DOI: 10.1038/nmeth.3899
Nanoscale imaging of RNA with expansion microscopy
Abstract
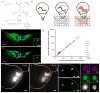
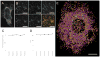
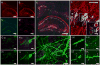
The ability to image RNA identity and location with nanoscale precision in intact tissues is of great interest for defining cell types and states in normal and pathological biological settings. Here, we present a strategy for expansion microscopy of RNA. We developed a small-molecule linker that enables RNA to be covalently attached to a swellable polyelectrolyte gel synthesized throughout a biological specimen. Then, postexpansion, fluorescent in situ hybridization (FISH) imaging of RNA can be performed with high yield and specificity as well as single-molecule precision in both cultured cells and intact brain tissue. Expansion FISH (ExFISH) separates RNAs and supports amplification of single-molecule signals (i.e., via hybridization chain reaction) as well as multiplexed RNA FISH readout. ExFISH thus enables super-resolution imaging of RNA structure and location with diffraction-limited microscopes in thick specimens, such as intact brain tissue and other tissues of importance to biology and medicine.
Conflict of interest statement
F.C., A.T.W., S. Alon, E.R.D., J.-B. C., A.M., G.M.C., and E.S.B. are inventors on one or more patents or patent applications related to the technologies here discussed. E.S.B. is co-founder of Expansion Technologies, a company whose goal is to facilitate access to expansion microscopy technologies by the scientific community.
Figures
Comment in
-
The quest for multiplexed spatially resolved transcriptional profiling.Nat Methods. 2016 Jul 28;13(8):623-4. doi: 10.1038/nmeth.3924. Nat Methods. 2016. PMID: 27467724 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

