Altered proliferation and networks in neural cells derived from idiopathic autistic individuals
- PMID: 27378147
- PMCID: PMC5215991
- DOI: 10.1038/mp.2016.95
Altered proliferation and networks in neural cells derived from idiopathic autistic individuals
Abstract
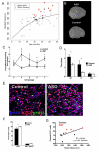
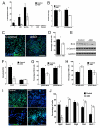
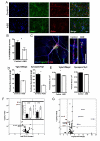
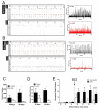
Autism spectrum disorders (ASD) are common, complex and heterogeneous neurodevelopmental disorders. Cellular and molecular mechanisms responsible for ASD pathogenesis have been proposed based on genetic studies, brain pathology and imaging, but a major impediment to testing ASD hypotheses is the lack of human cell models. Here, we reprogrammed fibroblasts to generate induced pluripotent stem cells, neural progenitor cells (NPCs) and neurons from ASD individuals with early brain overgrowth and non-ASD controls with normal brain size. ASD-derived NPCs display increased cell proliferation because of dysregulation of a β-catenin/BRN2 transcriptional cascade. ASD-derived neurons display abnormal neurogenesis and reduced synaptogenesis leading to functional defects in neuronal networks. Interestingly, defects in neuronal networks could be rescued by insulin growth factor 1 (IGF-1), a drug that is currently in clinical trials for ASD. This work demonstrates that selection of ASD subjects based on endophenotypes unraveled biologically relevant pathway disruption and revealed a potential cellular mechanism for the therapeutic effect of IGF-1.
Figures
References
-
- Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. The American journal of psychiatry. 1997;154(2):185–190. - PubMed
-
- Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45(6):691–699. - PubMed
-
- Garber K. Neuroscience. Autism’s cause may reside in abnormalities at the synapse. Science. 2007;317(5835):190–191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

