A highly diverse, desert-like microbial biocenosis on solar panels in a Mediterranean city
- PMID: 27378552
- PMCID: PMC4932501
- DOI: 10.1038/srep29235
A highly diverse, desert-like microbial biocenosis on solar panels in a Mediterranean city
Abstract
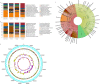
Microorganisms colonize a wide range of natural and artificial environments although there are hardly any data on the microbial ecology of one the most widespread man-made extreme structures: solar panels. Here we show that solar panels in a Mediterranean city (Valencia, Spain) harbor a highly diverse microbial community with more than 500 different species per panel, most of which belong to drought-, heat- and radiation-adapted bacterial genera, and sun-irradiation adapted epiphytic fungi. The taxonomic and functional profiles of this microbial community and the characterization of selected culturable bacteria reveal the existence of a diverse mesophilic microbial community on the panels' surface. This biocenosis proved to be more similar to the ones inhabiting deserts than to any human or urban microbial ecosystem. This unique microbial community shows different day/night proteomic profiles; it is dominated by reddish pigment- and sphingolipid-producers, and is adapted to withstand circadian cycles of high temperatures, desiccation and solar radiation.
Figures

References
-
- Corliss J. B. et al. Submarine thermal sprirngs on the galapagos rift. Science 203, 1073–1083 (1979). - PubMed
-
- Singh A. & Lal R. Sphingobium ummariense sp. nov., a hexachlorocyclohexane (HCH)-degrading bacterium, isolated from HCH-contaminated soil. Int. J. Syst. Evol. Microbiol. 59, 162–166 (2009). - PubMed
-
- Neilson J. W. et al. Life at the hyperarid margin: novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 16, 553–566 (2012). - PubMed
-
- Rosche B., Li X. Z., Hauer B., Schmid A. & Buehler K. Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 27, 636–643 (2009). - PubMed
-
- Williamson A. J. et al. Microbial reduction of U(VI) under alkaline conditions: implications for radioactive waste geodisposal. Environ. Sci. Technol. 48, 13549–13556 (2014). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

