Identification of consensus binding sites clarifies FMRP binding determinants
- PMID: 27378784
- PMCID: PMC5001617
- DOI: 10.1093/nar/gkw593
Identification of consensus binding sites clarifies FMRP binding determinants
Abstract
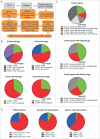
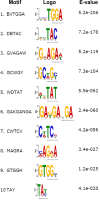
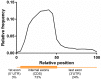
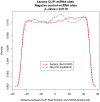
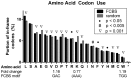
Fragile X mental retardation protein (FMRP) is a multifunctional RNA-binding protein with crucial roles in neuronal development and function. Efforts aimed at elucidating how FMRP target mRNAs are selected have produced divergent sets of target mRNA and putative FMRP-bound motifs, and a clear understanding of FMRP's binding determinants has been lacking. To clarify FMRP's binding to its target mRNAs, we produced a shared dataset of FMRP consensus binding sequences (FCBS), which were reproducibly identified in two published FMRP CLIP sequencing datasets. This comparative dataset revealed that of the various sequence and structural motifs that have been proposed to specify FMRP binding, the short sequence motifs TGGA and GAC were corroborated, and a novel TAY motif was identified. In addition, the distribution of the FCBS set demonstrates that FMRP preferentially binds to the coding region of its targets but also revealed binding along 3' UTRs in a subset of target mRNAs. Beyond probing these putative motifs, the FCBS dataset of reproducibly identified FMRP binding sites is a valuable tool for investigating FMRP targets and function.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. - PubMed
-
- Chen L., Yun S.W., Seto J., Liu W., Toth M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. - PubMed
-
- Dolzhanskaya N., Sung Y.J., Conti J., Currie J.R., Denman R.B. The fragile X mental retardation protein interacts with U-rich RNAs in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 2003;305:434–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

