Structural Studies of GABAA Receptor Binding Sites: Which Experimental Structure Tells us What?
- PMID: 27378845
- PMCID: PMC4910578
- DOI: 10.3389/fnmol.2016.00044
Structural Studies of GABAA Receptor Binding Sites: Which Experimental Structure Tells us What?
Abstract
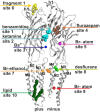
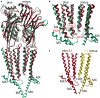
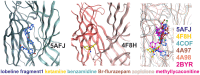
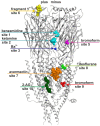
Atomic resolution structures of cys-loop receptors, including one of a γ-aminobutyric acid type A receptor (GABAA receptor) subtype, allow amazing insights into the structural features and conformational changes that these pentameric ligand-gated ion channels (pLGICs) display. Here we present a comprehensive analysis of more than 30 cys-loop receptor structures of homologous proteins that revealed several allosteric binding sites not previously described in GABAA receptors. These novel binding sites were examined in GABAA receptor homology models and assessed as putative candidate sites for allosteric ligands. Four so far undescribed putative ligand binding sites were proposed for follow up studies based on their presence in the GABAA receptor homology models. A comprehensive analysis of conserved structural features in GABAA and glycine receptors (GlyRs), the glutamate gated ion channel, the bacterial homologs Erwinia chrysanthemi (ELIC) and Gloeobacter violaceus GLIC, and the serotonin type 3 (5-HT3) receptor was performed. The conserved features were integrated into a master alignment that led to improved homology models. The large fragment of the intracellular domain that is present in the structure of the 5-HT3 receptor was utilized to generate GABAA receptor models with a corresponding intracellular domain fragment. Results of mutational and photoaffinity ligand studies in GABAA receptors were analyzed in the light of the model structures. This led to an assignment of candidate ligands to two proposed novel pockets, candidate binding sites for furosemide and neurosteroids in the trans-membrane domain were identified. The homology models can serve as hypotheses generators, and some previously controversial structural interpretations of biochemical data can be resolved in the light of the presented multi-template approach to comparative modeling. Crystal and cryo-EM microscopic structures of the closest homologs that were solved in different conformational states provided important insights into structural rearrangements of binding sites during conformational transitions. The impact of structural variation and conformational motion on the shape of the investigated binding sites was analyzed. Rules for best template and alignment choice were obtained and can generally be applied to modeling of cys-loop receptors. Overall, we provide an updated structure based view of ligand binding sites present in GABAA receptors.
Keywords: GABAA receptors; allosteric modulatory sites; binding pockets; conformations; subtype selectivity.
Figures



References
-
- Araujo F., Ruano D., Vitorica J. (1999). Native γ-aminobutyric acid type A receptors from rat hippocampus, containing both α 1 and α 5 subunits, exhibit a single benzodiazepine binding site with α 5 pharmacological properties. J. Pharmacol. Exp. Ther. 290, 989–997. Available online at: http://www.ncbi.nlm.nih.gov/pubmed/10454469 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

