Combined Linkage and Association Mapping Reveals QTL and Candidate Genes for Plant and Ear Height in Maize
- PMID: 27379126
- PMCID: PMC4908132
- DOI: 10.3389/fpls.2016.00833
Combined Linkage and Association Mapping Reveals QTL and Candidate Genes for Plant and Ear Height in Maize
Abstract
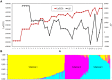
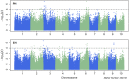
Plant height (PH) and ear height (EH) are two very important agronomic traits related to the population density and lodging in maize. In order to better understand of the genetic basis of nature variation in PH and EH, two bi-parental populations and one genome-wide association study (GWAS) population were used to map quantitative trait loci (QTL) for both traits. Phenotypic data analysis revealed a wide normal distribution and high heritability for PH and EH in the three populations, which indicated that maize height is a highly polygenic trait. A total of 21 QTL for PH and EH in three common genomic regions (bin 1.05, 5.04/05, and 6.04/05) were identified by QTL mapping in the two bi-parental populations under multiple environments. Additionally, 41 single nucleotide polymorphisms (SNPs) were identified for PH and EH by GWAS, of which 29 SNPs were located in 19 unique candidate gene regions. Most of the candidate genes were related to plant growth and development. One QTL on Chromosome 1 was further verified in a near-isogenic line (NIL) population, and GWAS identified a C2H2 zinc finger family protein that maybe the candidate gene for this QTL. These results revealed that nature variation of PH and EH are strongly controlled by multiple genes with low effect and facilitated a better understanding of the underlying mechanism of height in maize.
Keywords: association mapping; candidate gene; ear height; maize; plant height; quantitative trait locus.
Figures


References
-
- Agrama H. A. S., Moussa M. E. (1996). Mapping QTLs in breeding for drought tolerance in maize (Zea mays L.). Euphytica 91, 89–97. 10.1007/BF00035278 - DOI
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources

