Signal transduction controls heterogeneous NF-κB dynamics and target gene expression through cytokine-specific refractory states
- PMID: 27381163
- PMCID: PMC4935804
- DOI: 10.1038/ncomms12057
Signal transduction controls heterogeneous NF-κB dynamics and target gene expression through cytokine-specific refractory states
Abstract
Cells respond dynamically to pulsatile cytokine stimulation. Here we report that single, or well-spaced pulses of TNFα (>100 min apart) give a high probability of NF-κB activation. However, fewer cells respond to shorter pulse intervals (<100 min) suggesting a heterogeneous refractory state. This refractory state is established in the signal transduction network downstream of TNFR and upstream of IKK, and depends on the level of the NF-κB system negative feedback protein A20. If a second pulse within the refractory phase is IL-1β instead of TNFα, all of the cells respond. This suggests a mechanism by which two cytokines can synergistically activate an inflammatory response. Gene expression analyses show strong correlation between the cellular dynamic response and NF-κB-dependent target gene activation. These data suggest that refractory states in the NF-κB system constitute an inherent design motif of the inflammatory response and we suggest that this may avoid harmful homogenous cellular activation.
Figures
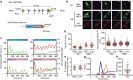
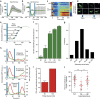
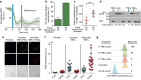

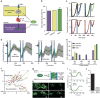
References
-
- Elowitz M. B., Levine A. J., Siggia E. D. & Swain P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0500346/MRC_/Medical Research Council/United Kingdom
- MR/K015885/1/MRC_/Medical Research Council/United Kingdom
- BB/K003097/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M008908/1/MRC_/Medical Research Council/United Kingdom
- BB/F005938/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

