Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax
- PMID: 27381764
- PMCID: PMC5205579
- DOI: 10.1016/j.ijpara.2016.05.008
Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax
Abstract
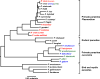
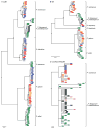
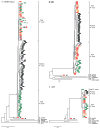
Plasmodium falciparum and Plasmodium vivax account for more than 95% of all human malaria infections, and thus pose a serious public health challenge. To control and potentially eliminate these pathogens, it is important to understand their origins and evolutionary history. Until recently, it was widely believed that P. falciparum had co-evolved with humans (and our ancestors) over millions of years, whilst P. vivax was assumed to have emerged in southeastern Asia following the cross-species transmission of a parasite from a macaque. However, the discovery of a multitude of Plasmodium spp. in chimpanzees and gorillas has refuted these theories and instead revealed that both P. falciparum and P. vivax evolved from parasites infecting wild-living African apes. It is now clear that P. falciparum resulted from a recent cross-species transmission of a parasite from a gorilla, whilst P. vivax emerged from an ancestral stock of parasites that infected chimpanzees, gorillas and humans in Africa, until the spread of the protective Duffy-negative mutation eliminated P. vivax from human populations there. Although many questions remain concerning the biology and zoonotic potential of the P. falciparum- and P. vivax-like parasites infecting apes, comparative genomics, coupled with functional parasite and vector studies, are likely to yield new insights into ape Plasmodium transmission and pathogenesis that are relevant to the treatment and prevention of human malaria.
Keywords: African apes; Evolution; Laverania; Malaria; Plasmodium falciparum; Plasmodium vivax; Zoonotic transmission.
Copyright © 2016 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Amambua-Ngwa A, Tetteh KK, Manske M, Gomez-Escobar N, Stewart LB, Deerhake ME, Cheeseman IH, Newbold CI, Holder AA, Knuepfer E, Janha O, Jallow M, Campino S, Macinnis B, Kwiatkowski DP, Conway DJ. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8:e1002992. - PMC - PubMed
-
- Ayouba A, Mouacha F, Learn GH, Mpoudi-Ngole E, Rayner JC, Sharp PM, Hahn BH, Delaporte E, Peeters M. Ubiquitous Hepatocystis infections, but no evidence of Plasmodium falciparum-like malaria parasites in wild greater spot-nosed monkeys (Cercopithecus nictitans) Int J Parasitol. 2012;42:709–713. - PMC - PubMed
-
- Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, Battle KE, Moyes CL, Henry A, Eckhoff PA, Wenger EA, Briet O, Penny MA, Smith TA, Bennett A, Yukich J, Eisele TP, Griffin JT, Fergus CA, Lynch M, Lindgren F, Cohen JM, Murray CL, Smith DL, Hay SI, Cibulskis RE, Gething PW. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–211. - PMC - PubMed
-
- Blacklock B, Adler S. A parasite resembling Plasmodium falciparum in a chimpanzee. Ann Trop Med Parasitol. 1922;XVI:99–106.
-
- Bopp SER, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, Fidock DA, Denchi EL, Winzeler EA. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9:e1003293. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

