HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-κB pathway in a transcription-independent manner
- PMID: 27382069
- PMCID: PMC5009750
- DOI: 10.1093/nar/gkw606
HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-κB pathway in a transcription-independent manner
Abstract
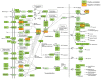
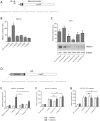
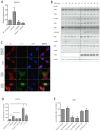
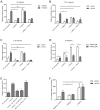
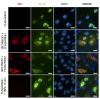
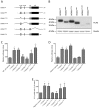
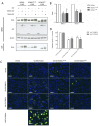
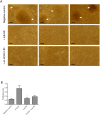
HOX proteins define a family of key transcription factors regulating animal embryogenesis. HOX genes have also been linked to oncogenesis and HOXA1 has been described to be active in several cancers, including breast cancer. Through a proteome-wide interaction screening, we previously identified the TNFR-associated proteins RBCK1/HOIL-1 and TRAF2 as HOXA1 interactors suggesting that HOXA1 is functionally linked to the TNF/NF-κB signaling pathway. Here, we reveal a strong positive correlation between expression of HOXA1 and of members of the TNF/NF-κB pathway in breast tumor datasets. Functionally, we demonstrate that HOXA1 can activate NF-κB and operates upstream of the NF-κB inhibitor IκB. Consistently, we next demonstrate that the HOXA1-mediated activation of NF-κB is non-transcriptional and that RBCK1 and TRAF2 influences on NF-κB are epistatic to HOXA1. We also identify an 11 Histidine repeat and the homeodomain of HOXA1 to be required both for RBCK1 and TRAF2 interaction and NF-κB stimulation. Finally, we highlight that activation of NF-κB is crucial for HOXA1 oncogenic activity.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Rezsohazy R., Saurin A.J., Maurel-Zaffran C., Graba Y. Cellular and molecular insights into Hox protein action. Development. 2015;142:1212–1227. - PubMed
-
- Alexander T., Nolte C., Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 2009;25:431–456. - PubMed
-
- Chisaka O., Musci T.S., Capecchi M.R. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992;355:516–520. - PubMed
-
- Mark M., Lufkin T., Vonesch J.L., Ruberte E., Olivo J.C., Dolle P., Gorry P., Lumsden A., Chambon P. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119:319–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

