Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds
- PMID: 27382413
- PMCID: PMC4932711
- DOI: 10.1186/s13068-016-0555-5
Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds
Abstract
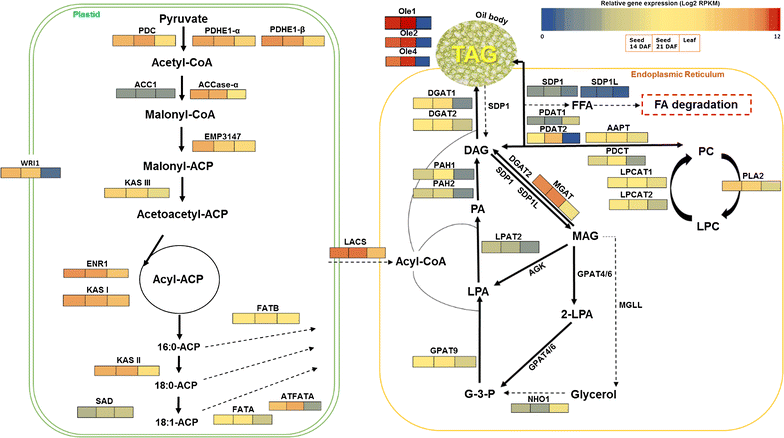
Background: Camelina sativa is an emerging dedicated oilseed crop designed for biofuel and biodiesel applications as well as a source for edible and general-purpose oils. Such valuable oilseed crop is subjected to plant breeding programs and is suggested for large-scale production of better seed and oil quality. To accomplish this objective and to further enhance its oil content, a better understanding of lipid metabolism at the molecular level in this plant is critical. Here, we applied tissue transcriptomics and lipid composition analysis to identify and profile the genes and gene networks associated with triacylglycerol (TAG) biosynthesis, and to investigate how those genes are interacting to determine the quantity and quality of Camelina oil during seed development.
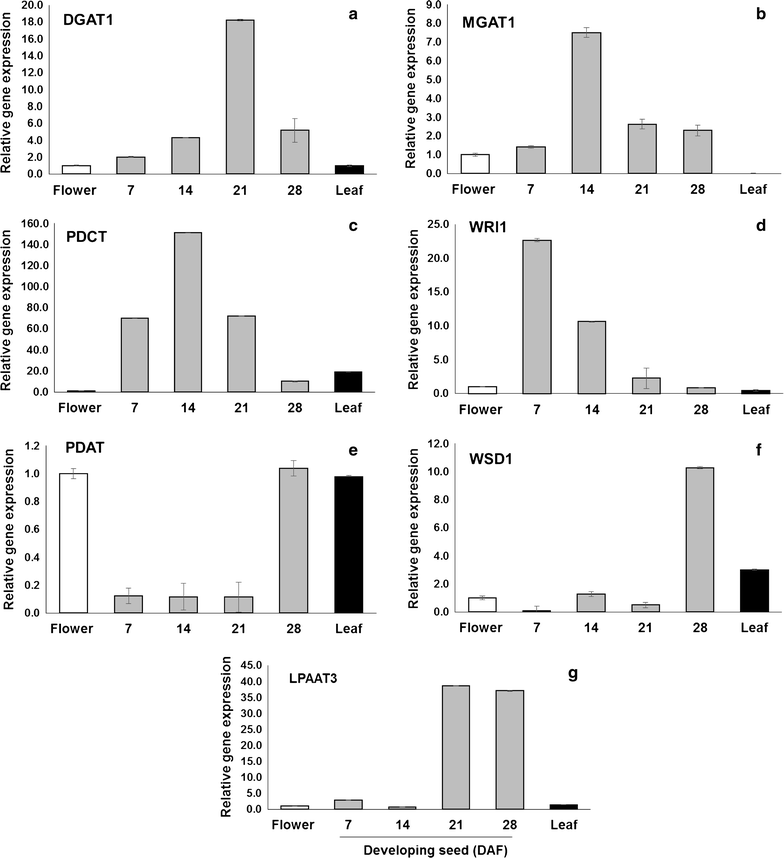
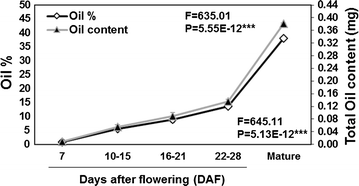
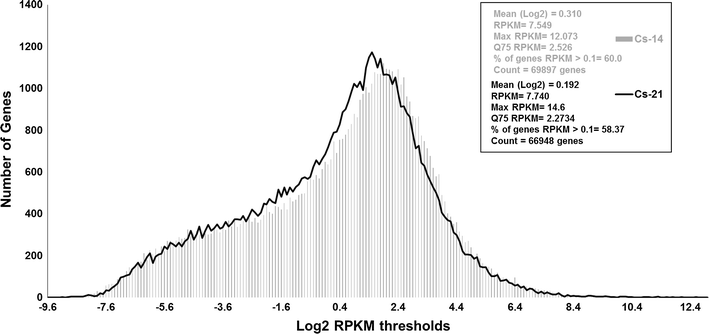
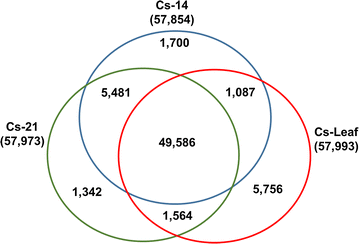
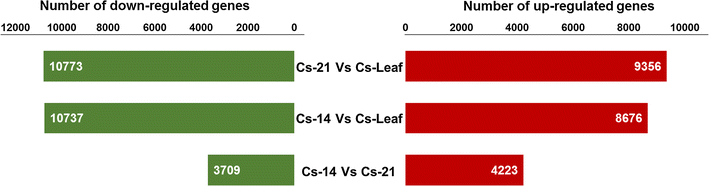
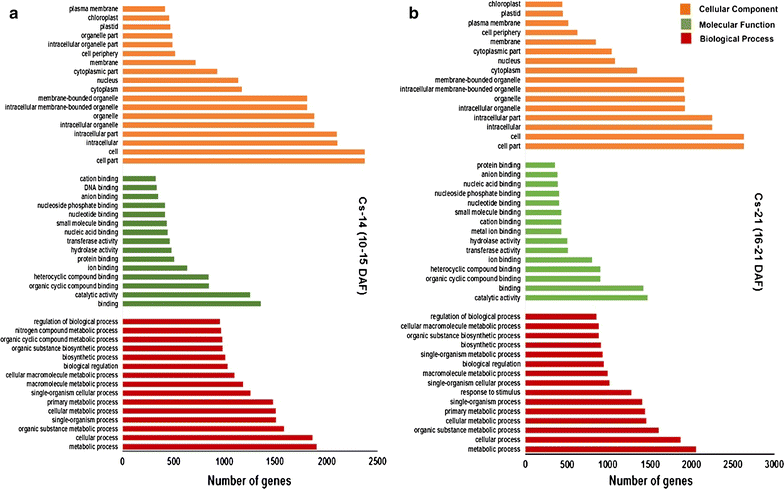
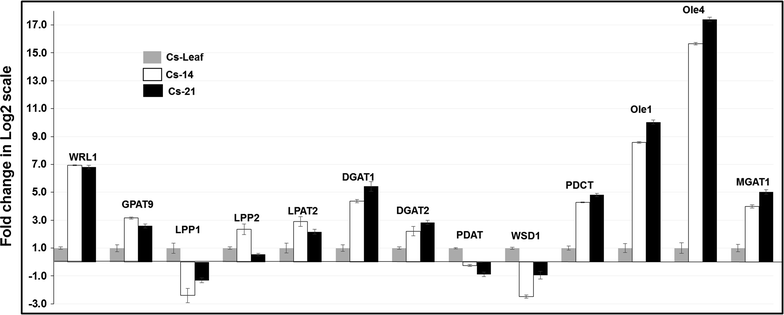
Results: Our Camelina transcriptome data analysis revealed an approximate of 57,854 and 57,973 genes actively expressing in developing seeds (RPKM ≥ 0.1) at 10-15 (Cs-14) and 16-21 (Cs-21) days after flowering (DAF), respectively. Of these, 7932 genes showed temporal and differential gene expression during the seed development (log2 fold change ≥1.5 or ≤-1.5; P ≤ 0.05). The differentially expressed genes (DEGs) were annotated and were found to be involved in distinct functional categories and metabolic pathways. Furthermore, performing quantitative real-time PCR for selected candidate genes associated with TAG biosynthesis validated RNA-seq data. Our results showed strong positive correlations between the expression abundance measured using both qPCR and RNA-Seq technologies. Furthermore, the analysis of fatty-acid content and composition revealed major changes throughout seed development, with the amount of oil accumulate rapidly at early mid seed development stages (from 16-28 DAF onwards), while no important changes were observed in the fatty-acid profile between seeds at 28 DAF and mature seeds.
Conclusions: This study is highly useful for understanding the regulation of TAG biosynthesis and identifying the rate-limiting steps in TAG pathways at seed development stages, providing a precise selection of candidate genes for developing Camelina varieties with improved seed and oil yields.
Keywords: Camelina sativa; Fatty-acid profiling; Lipid metabolism; Transcriptome profiling; Triacylglycerol biosynthesis.
Figures
References
-
- Kennedy EP. Biosynthesis of complex lipids. Federation Proceedings. 1961;20:934–940. - PubMed
-
- McVay KA, Lamb PF. Camelina production in Montana. Montana State University. 2008. http://store.msuextension.org/publications/AgandNaturalResources/MT20070....
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

