Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores
- PMID: 27383986
- PMCID: PMC5539988
- DOI: 10.1038/nature18629
Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores
Abstract
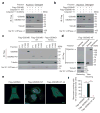
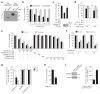
Inflammatory caspases (caspases 1, 4, 5 and 11) are activated in response to microbial infection and danger signals. When activated, they cleave mouse and human gasdermin D (GSDMD) after Asp276 and Asp275, respectively, to generate an N-terminal cleavage product (GSDMD-NT) that triggers inflammatory death (pyroptosis) and release of inflammatory cytokines such as interleukin-1β. Cleavage removes the C-terminal fragment (GSDMD-CT), which is thought to fold back on GSDMD-NT to inhibit its activation. However, how GSDMD-NT causes cell death is unknown. Here we show that GSDMD-NT oligomerizes in membranes to form pores that are visible by electron microscopy. GSDMD-NT binds to phosphatidylinositol phosphates and phosphatidylserine (restricted to the cell membrane inner leaflet) and cardiolipin (present in the inner and outer leaflets of bacterial membranes). Mutation of four evolutionarily conserved basic residues blocks GSDMD-NT oligomerization, membrane binding, pore formation and pyroptosis. Because of its lipid-binding preferences, GSDMD-NT kills from within the cell, but does not harm neighbouring mammalian cells when it is released during pyroptosis. GSDMD-NT also kills cell-free bacteria in vitro and may have a direct bactericidal effect within the cytosol of host cells, but the importance of direct bacterial killing in controlling in vivo infection remains to be determined.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. - PubMed
-
- Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. - PubMed
-
- Law RH, et al. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

