CENPT bridges adjacent CENPA nucleosomes on young human α-satellite dimers
- PMID: 27384170
- PMCID: PMC5052034
- DOI: 10.1101/gr.204784.116
CENPT bridges adjacent CENPA nucleosomes on young human α-satellite dimers
Abstract
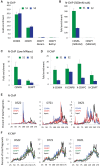
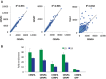
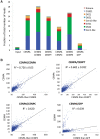
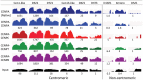
Nucleosomes containing the CenH3 (CENPA or CENP-A) histone variant replace H3 nucleosomes at centromeres to provide a foundation for kinetochore assembly. CENPA nucleosomes are part of the constitutive centromere associated network (CCAN) that forms the inner kinetochore on which outer kinetochore proteins assemble. Two components of the CCAN, CENPC and the histone-fold protein CENPT, provide independent connections from the ∼171-bp centromeric α-satellite repeat units to the outer kinetochore. However, the spatial relationship between CENPA nucleosomes and these two branches remains unclear. To address this issue, we use a base-pair resolution genomic readout of protein-protein interactions, comparative chromatin immunoprecipitation (ChIP) with sequencing, together with sequential ChIP, to infer the in vivo molecular architecture of the human CCAN. In contrast to the currently accepted model in which CENPT associates with H3 nucleosomes, we find that CENPT is centered over the CENPB box between two well-positioned CENPA nucleosomes on the most abundant centromeric young α-satellite dimers and interacts with the CENPB/CENPC complex. Upon cross-linking, the entire CENPA/CENPB/CENPC/CENPT complex is nuclease-protected over an α-satellite dimer that comprises the fundamental unit of centromeric chromatin. We conclude that CENPA/CENPC and CENPT pathways for kinetochore assembly are physically integrated over young α-satellite dimers.
© 2016 Thakur and Henikoff; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Conserved and divergent mechanisms of inner kinetochore assembly onto centromeric chromatin.Curr Opin Struct Biol. 2023 Aug;81:102638. doi: 10.1016/j.sbi.2023.102638. Epub 2023 Jun 20. Curr Opin Struct Biol. 2023. PMID: 37343495 Review.
-
Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points.J Cell Biol. 2017 Mar 6;216(3):607-621. doi: 10.1083/jcb.201608083. Epub 2017 Feb 24. J Cell Biol. 2017. PMID: 28235947 Free PMC article.
-
Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice.Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):1148-53. doi: 10.1073/pnas.97.3.1148. Proc Natl Acad Sci U S A. 2000. PMID: 10655499 Free PMC article.
-
CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore.Cell. 2008 Dec 12;135(6):1039-52. doi: 10.1016/j.cell.2008.10.019. Cell. 2008. PMID: 19070575
-
The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle.Open Biol. 2020 Jun;10(6):200051. doi: 10.1098/rsob.200051. Epub 2020 Jun 10. Open Biol. 2020. PMID: 32516549 Free PMC article. Review.
Cited by
-
Mapping separase-mediated cleavage in situ.NAR Genom Bioinform. 2022 Nov 18;4(4):lqac085. doi: 10.1093/nargab/lqac085. eCollection 2022 Dec. NAR Genom Bioinform. 2022. PMID: 36415827 Free PMC article.
-
Unexpected conformational variations of the human centromeric chromatin complex.Genes Dev. 2018 Jan 1;32(1):20-25. doi: 10.1101/gad.307736.117. Epub 2018 Jan 31. Genes Dev. 2018. PMID: 29386331 Free PMC article.
-
Classification and monomer-by-monomer annotation dataset of suprachromosomal family 1 alpha satellite higher-order repeats in hg38 human genome assembly.Data Brief. 2019 Mar 8;24:103708. doi: 10.1016/j.dib.2019.103708. eCollection 2019 Jun. Data Brief. 2019. PMID: 30989093 Free PMC article.
-
CENP-C/H/I/K/M/T/W/N/L and hMis12 but not CENP-S/X participate in complex formation in the nucleoplasm of living human interphase cells outside centromeres.PLoS One. 2018 Mar 6;13(3):e0192572. doi: 10.1371/journal.pone.0192572. eCollection 2018. PLoS One. 2018. PMID: 29509805 Free PMC article.
-
Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80.Mol Cell. 2018 Sep 20;71(6):923-939.e10. doi: 10.1016/j.molcel.2018.07.038. Epub 2018 Aug 30. Mol Cell. 2018. PMID: 30174292 Free PMC article.
References
-
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y. 2001. α-Satellite DNA of primates: old and new families. Chromosoma 110: 253–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
