Genetic Lineage and Reassortment of Influenza C Viruses Circulating between 1947 and 2014
- PMID: 27384661
- PMCID: PMC5008092
- DOI: 10.1128/JVI.00969-16
Genetic Lineage and Reassortment of Influenza C Viruses Circulating between 1947 and 2014
Abstract
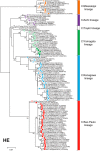
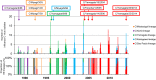
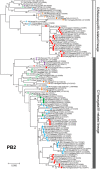
Since influenza C virus was first isolated in 1947, the virus has been only occasionally isolated by cell culture; there are only four strains for which complete genome sequences are registered. Here, we analyzed a total of 106 complete genomes, ranging from the first isolate from 1947 to recent isolates from 2014, to determine the genetic lineages of influenza C virus, the reassortment events, and the rates of nucleotide substitution. The results showed that there are six lineages, named C/Taylor, C/Mississippi, C/Aichi, C/Yamagata, C/Kanagawa, and C/Sao Paulo. They contain both antigenic and genetic lineages of the hemagglutinin-esterase (HE) gene, and the internal genes PB2, PB1, P3, NP, M, and NS are divided into two major lineages, a C/Mississippi/80-related lineage and a C/Yamagata/81-related lineage. Reassortment events were found over the entire period of 68 years. Several outbreaks of influenza C virus between 1990 and 2014 in Japan consisted of reassortant viruses, suggesting that the genomic constellation is related to influenza C virus epidemics. The nucleotide sequences were highly homologous to each other. The minimum percent identity between viruses ranged from 91.1% for the HE gene to 96.1% for the M gene, and the rate of nucleotide substitution for the HE gene was the highest, at 5.20 × 10(-4) substitutions/site/year. These results indicate that reassortment is an important factor that increases the genetic diversity of influenza C virus, resulting in its ability to prevail in humans. IMPORTANCE Influenza C virus is a pathogen that causes acute respiratory illness in children and results in hospitalization of infants. We previously demonstrated (Y. Matsuzaki et al., J Clin Virol 61:87-93, 2014, http://dx.doi.org/10.1016/j.jcv.2014.06.017) that periodic epidemics of this virus occurred in Japan between 1996 and 2014 and that replacement of the dominant antigenic group occurred every several years as a result of selection by herd immunity. However, the antigenicity of the HE glycoprotein is highly stable, and antigenic drift has not occurred for at least 30 years. Here, we analyzed a total of 106 complete genomes spanning 68 years for the first time, and we found that influenza C viruses are circulating worldwide while undergoing reassortment as well as selection by herd immunity, resulting in an increased ability to prevail in humans. The results presented in this study contribute to the understanding of the evolution, including reassortment events, underlying influenza C virus epidemics.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

