Tumour sampling method can significantly influence gene expression profiles derived from neoadjuvant window studies
- PMID: 27384960
- PMCID: PMC4935948
- DOI: 10.1038/srep29434
Tumour sampling method can significantly influence gene expression profiles derived from neoadjuvant window studies
Abstract
Patient-matched transcriptomic studies using tumour samples before and after treatment allow inter-patient heterogeneity to be controlled, but tend not to include an untreated comparison. Here, Illumina BeadArray technology was used to measure dynamic changes in gene expression from thirty-seven paired diagnostic core and surgically excised breast cancer biopsies obtained from women receiving no treatment prior to surgery, to determine the impact of sampling method and tumour heterogeneity. Despite a lack of treatment and perhaps surprisingly, consistent changes in gene expression were identified during the diagnosis-surgery interval (48 up, 2 down; Siggenes FDR 0.05) in a manner independent of both subtype and sampling-interval length. Instead, tumour sampling method was seen to directly impact gene expression, with similar effects additionally identified in six published breast cancer datasets. In contrast with previous findings, our data does not support the concept of a significant wounding or immune response following biopsy in the absence of treatment and instead implicates a hypoxic response following the surgical biopsy. Whilst sampling-related gene expression changes are evident in treated samples, they are secondary to those associated with response to treatment. Nonetheless, sampling method remains a potential confounding factor for neoadjuvant study design.
Figures

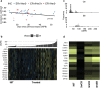
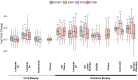
References
-
- Dekker T. J. A. et al. Reliability of core needle biopsy for determining ER and HER2 status in breast cancer. Ann. Oncol. 24, 931–937 (2013). - PubMed
-
- van de Vijver M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002). - PubMed
-
- Filipits M. et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 17, 6012–6020 (2011). - PubMed
-
- Ma X.-J. et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin. Cancer Res. 14, 2601–2608 (2008). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

