Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities
- PMID: 27385827
- PMCID: PMC4961173
- DOI: 10.1073/pnas.1600974113
Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities
Abstract
Horizontal gene transfer is a fundamental process in bacterial evolution that can accelerate adaptation via the sharing of genes between lineages. Conjugative plasmids are the principal genetic elements mediating the horizontal transfer of genes, both within and between bacterial species. In some species, plasmids are unstable and likely to be lost through purifying selection, but when alternative hosts are available, interspecific plasmid transfer could counteract this and maintain access to plasmid-borne genes. To investigate the evolutionary importance of alternative hosts to plasmid population dynamics in an ecologically relevant environment, we established simple soil microcosm communities comprising two species of common soil bacteria, Pseudomonas fluorescens and Pseudomonas putida, and a mercury resistance (Hg(R)) plasmid, pQBR57, both with and without positive selection [i.e., addition of Hg(II)]. In single-species populations, plasmid stability varied between species: although pQBR57 survived both with and without positive selection in P. fluorescens, it was lost or replaced by nontransferable Hg(R) captured to the chromosome in P. putida A simple mathematical model suggests these differences were likely due to pQBR57's lower intraspecific conjugation rate in P. putida By contrast, in two-species communities, both models and experiments show that interspecific conjugation from P. fluorescens allowed pQBR57 to persist in P. putida via source-sink transfer dynamics. Moreover, the replacement of pQBR57 by nontransferable chromosomal Hg(R) in P. putida was slowed in coculture. Interspecific transfer allows plasmid survival in host species unable to sustain the plasmid in monoculture, promoting community-wide access to the plasmid-borne accessory gene pool and thus potentiating future evolvability.
Keywords: horizontal gene transfer; microbial ecology; mobile genetic elements; plasmids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
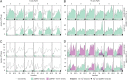
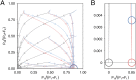
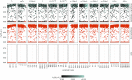


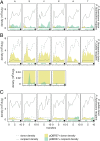
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

