Reduced evolutionary rate in reemerged Ebola virus transmission chains
- PMID: 27386513
- PMCID: PMC4928956
- DOI: 10.1126/sciadv.1600378
Reduced evolutionary rate in reemerged Ebola virus transmission chains
Abstract
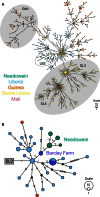
On 29 June 2015, Liberia's respite from Ebola virus disease (EVD) was interrupted for the second time by a renewed outbreak ("flare-up") of seven confirmed cases. We demonstrate that, similar to the March 2015 flare-up associated with sexual transmission, this new flare-up was a reemergence of a Liberian transmission chain originating from a persistently infected source rather than a reintroduction from a reservoir or a neighboring country with active transmission. Although distinct, Ebola virus (EBOV) genomes from both flare-ups exhibit significantly low genetic divergence, indicating a reduced rate of EBOV evolution during persistent infection. Using this rate of change as a signature, we identified two additional EVD clusters that possibly arose from persistently infected sources. These findings highlight the risk of EVD flare-ups even after an outbreak is declared over.
Keywords: Ebola virus; Ebola virus disease; Liberia; Western Africa; flare-up; persistent infection; reduced evolutionary rate; reemerged; transmission chain.
Figures
References
-
- Mate S. E., Kugelman J. R., Nyenswah T. G., Ladner J. T., Wiley M. R., Cordier-Lassalle T., Christie A., Schroth G. P., Gross S. M., Davies-Wayne G. J., Shinde S. A., Murugan R., Sieh S. B., Badio M., Fakoli L., Taweh F., de Wit E., van Doremalen N., Munster V. J., Pettitt J., Prieto K., Humrighouse B. W., Ströher U., DiClaro J. W., Hensley L. E., Schoepp R. J., Safronetz D., Fair J., Kuhn J. H., Blackley D. J., Laney A. S., Williams D. E., Lo T., Gasasira A., Nichol S. T., Formenty P., Kateh F. N., De Cock K. M., Bolay F., Sanchez-Lockhart M., Palacios G., Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med. 373, 2448–2454 (2015). - PMC - PubMed
-
- Ladner J. T., Beitzel B., Chain P. S. G., Davenport M. G., Donaldson E. F., Frieman M., Kugelman J. R., Kuhn J. H., O’Rear J., Sabeti P. C., Wentworth D. E., Wiley M. R., Yu G.-Y.; Threat Characterization Consortium, Sozhamannan S., Bradburne C., Palacios G., Standards for sequencing viral genomes in the era of high-throughput sequencing. mBio 5, e01360-14 (2014). - PMC - PubMed
-
- Baize S., Pannetier D., Oestereich L., Rieger T., Koivogui L., Magassouba N., Soropogui B., Sow M. S., Keïta S., De Clerck H., Tiffany A., Dominguez G., Loua M., Traore A., Kolié M., Malano E. R., Heleze E., Bocquin A., Mely S., Raoul H., Caro V., Cadar D., Gabriel M., Pahlmann M., Tappe D., Schmidt-Chanasit J., Impouma B., Diallo A. K., Formenty P., Van Herp M., Günther S., Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 371, 1418–1425 (2014). - PubMed
-
- Gire S. K., Goba A., Andersen K. G., Sealfon R. S. G., Park D. J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G., Wohl S., Moses L. M., Yozwiak N. L., Winnicki S., Matranga C. B., Malboeuf C. M., Qu J., Gladden A. D., Schaffner S. F., Yang X., Jiang P. P., Nekoui M., Colubri A., Coomber M. R., Fonnie M., Moigboi A., Gbakie M., Kamara F. K., Tucker V., Konuwa E., Saffa S., Sellu J., Jalloh A. A., Kovoma A., Koninga J., Mustapha I., Kargbo K., Foday M., Yillah M., Kanneh F., Robert W., Massally J. L., Chapman S. B., Bochicchio J., Murphy C., Nusbaum C., Young S., Birren B. W., Grant D. S., Scheiffelin J. S., Lander E. S., Happi C., Gevao S. M., Gnirke A., Rambaut A., Garry R. F., Khan S. H., Sabeti P. C., Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345, 1369–1372 (2014). - PMC - PubMed
-
- Ladner J. T., Wiley M. R., Mate S., Dudas G., Prieto K., Lovett S., Nagle E. R., Beitzel B., Gilbert M. L., Fakoli L., Diclaro J. W. II, Schoepp R. J., Fair J., Kuhn J. H., Hensley L. E., Park D. J., Sabeti P. C., Rambaut A., Sanchez-Lockhart M., Bolay F. K., Kugelman J. R., Palacios G., Evolution and spread of Ebola virus in Liberia, 2014–2015. Cell Host Microbe 18, 659–669 (2015). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

