HIV-1 escapes from N332-directed antibody neutralization in an elite neutralizer by envelope glycoprotein elongation and introduction of unusual disulfide bonds
- PMID: 27388013
- PMCID: PMC4936165
- DOI: 10.1186/s12977-016-0279-4
HIV-1 escapes from N332-directed antibody neutralization in an elite neutralizer by envelope glycoprotein elongation and introduction of unusual disulfide bonds
Abstract
Background: Current HIV-1 immunogens are unable to induce antibodies that can neutralize a broad range of HIV-1 (broadly neutralizing antibodies; bNAbs). However, such antibodies are elicited in 10-30 % of HIV-1 infected individuals, and the co-evolution of the virus and the humoral immune responses in these individuals has attracted attention, because they can provide clues for vaccine design.
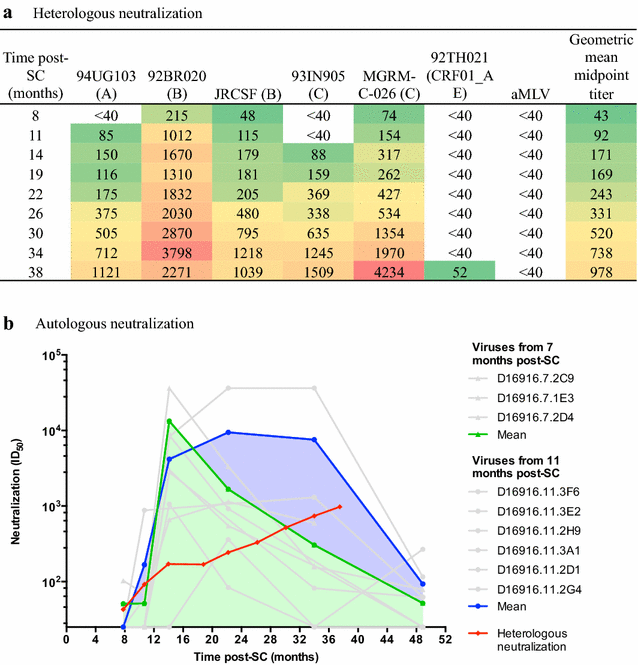
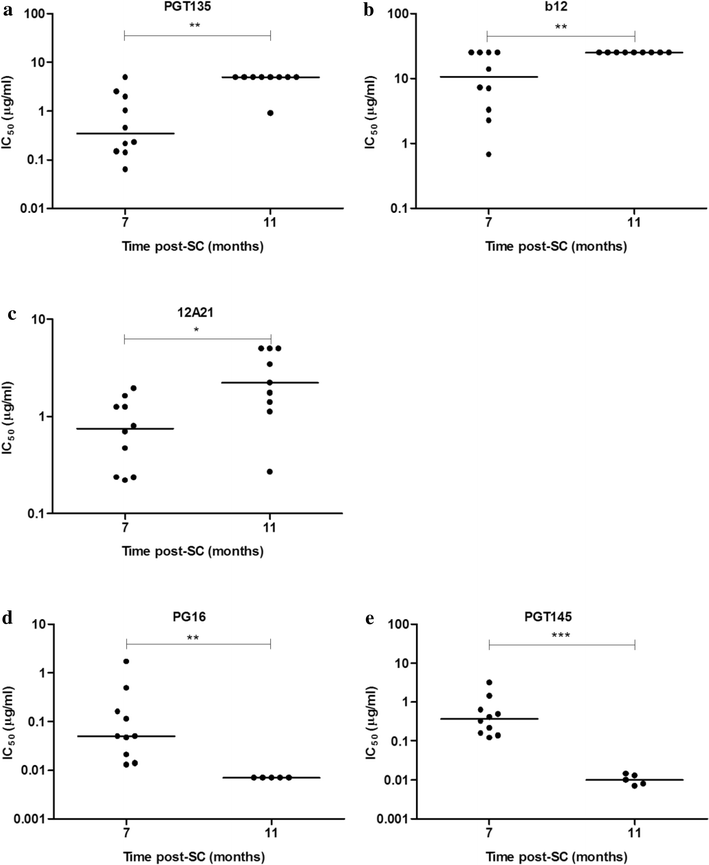
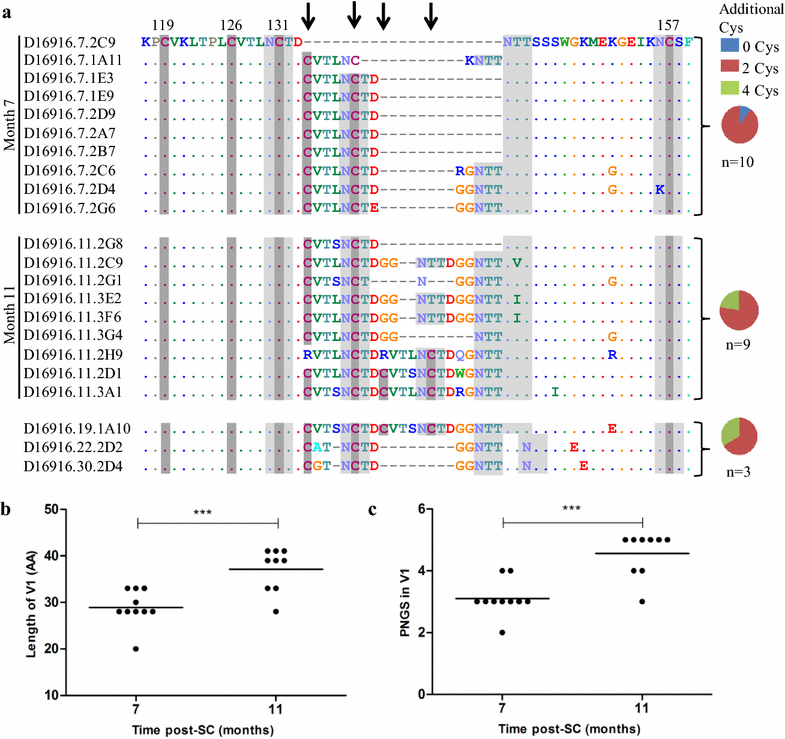
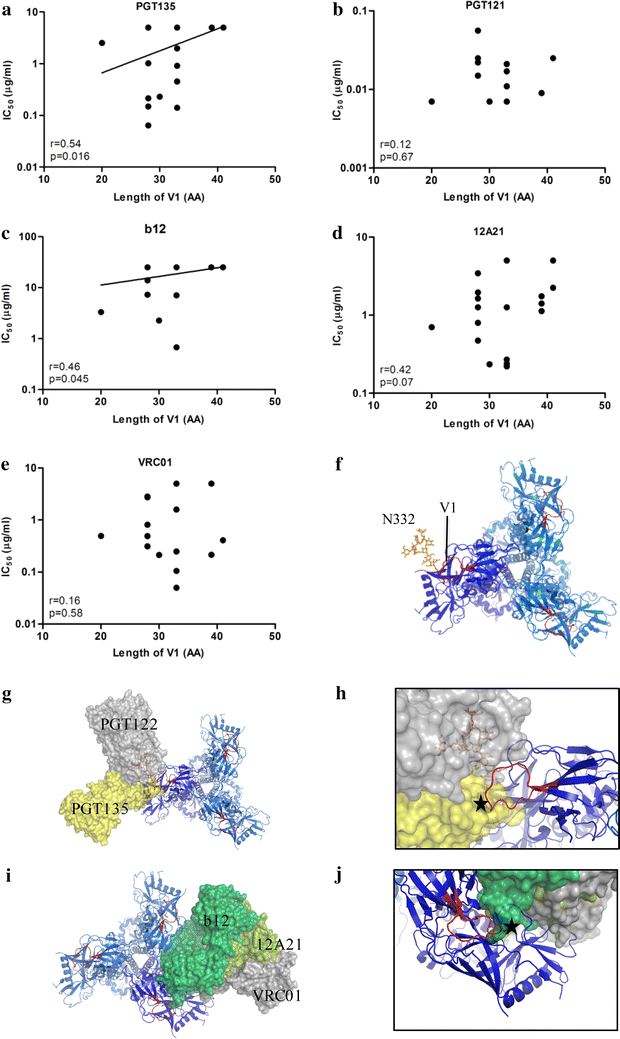
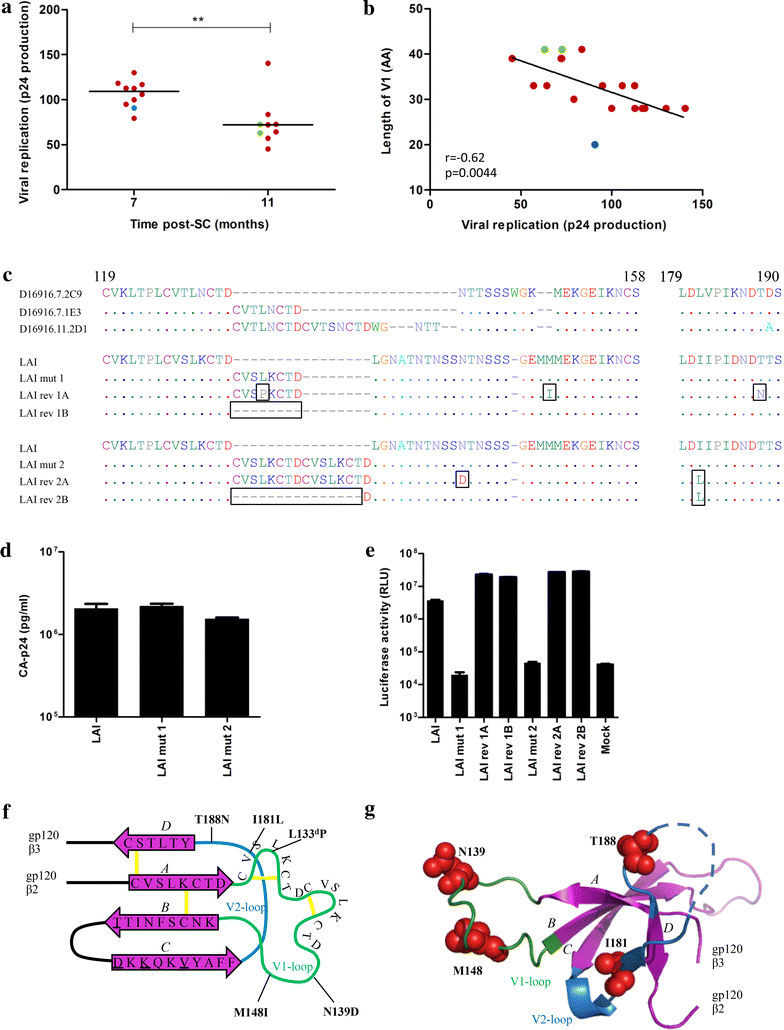
Results: Here we characterized the NAb responses and envelope glycoprotein evolution in an HIV-1 infected "elite neutralizer" of the Amsterdam Cohort Studies on HIV-1 infection and AIDS who developed an unusually potent bNAb response rapidly after infection. The NAb response was dependent on the N332-glycan and viral resistance against the N332-glycan dependent bNAb PGT135 developed over time but viral escape did not occur at or near this glycan. In contrast, the virus likely escaped by increasing V1 length, with up to 21 amino acids, accompanied by the introduction of 1-3 additional glycans, as well as 2-4 additional cysteine residues within V1.
Conclusions: In the individual studied here, HIV-1 escaped from N332-glycan directed NAb responses without changing the epitope itself, but by elongating a variable loop that shields this epitope.
Keywords: Broadly neutralizing antibodies; Cysteines; Envelope glycoprotein; Glycans; HIV-1; N332; Variable regions.
Figures
Similar articles
-
Cooperation between Strain-Specific and Broadly Neutralizing Responses Limited Viral Escape and Prolonged the Exposure of the Broadly Neutralizing Epitope.J Virol. 2017 Aug 24;91(18):e00828-17. doi: 10.1128/JVI.00828-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28679760 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
An HIV-1 Broadly Neutralizing Antibody from a Clade C-Infected Pediatric Elite Neutralizer Potently Neutralizes the Contemporaneous and Autologous Evolving Viruses.J Virol. 2019 Feb 5;93(4):e01495-18. doi: 10.1128/JVI.01495-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30429339 Free PMC article.
-
HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies.Expert Rev Vaccines. 2016;15(3):349-65. doi: 10.1586/14760584.2016.1129905. Epub 2016 Jan 8. Expert Rev Vaccines. 2016. PMID: 26654478 Review.
-
Beyond glycan barriers: non-cognate ligands and protein mimicry approaches to elicit broadly neutralizing antibodies for HIV-1.J Biomed Sci. 2024 Aug 21;31(1):83. doi: 10.1186/s12929-024-01073-y. J Biomed Sci. 2024. PMID: 39169357 Free PMC article. Review.
Cited by
-
Cooperation between Strain-Specific and Broadly Neutralizing Responses Limited Viral Escape and Prolonged the Exposure of the Broadly Neutralizing Epitope.J Virol. 2017 Aug 24;91(18):e00828-17. doi: 10.1128/JVI.00828-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28679760 Free PMC article.
-
HIV-1 clade C escapes broadly neutralizing autologous antibodies with N332 glycan specificity by distinct mechanisms.Retrovirology. 2016 Aug 30;13(1):60. doi: 10.1186/s12977-016-0297-2. Retrovirology. 2016. PMID: 27576440 Free PMC article.
-
Current Peptide and Protein Candidates Challenging HIV Therapy beyond the Vaccine Era.Viruses. 2017 Sep 29;9(10):281. doi: 10.3390/v9100281. Viruses. 2017. PMID: 28961190 Free PMC article. Review.
-
The expanding array of HIV broadly neutralizing antibodies.Retrovirology. 2018 Oct 16;15(1):70. doi: 10.1186/s12977-018-0453-y. Retrovirology. 2018. PMID: 30326938 Free PMC article. Review.
-
Envelope characteristics in individuals who developed neutralizing antibodies targeting different epitopes in HIV-1 subtype C infection.Virology. 2020 Jul;546:1-12. doi: 10.1016/j.virol.2020.03.003. Epub 2020 Mar 25. Virology. 2020. PMID: 32275203 Free PMC article.
References
-
- Doria-Rose NA, Klein RM, Manion MM, O’Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. - DOI - PMC - PubMed
-
- Euler Z, van den Kerkhof TL, van Gils MJ, Burger JA, Edo-Matas D, Phung P, Wrin T, Schuitemaker H. Longitudinal analysis of early HIV-1 specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. J Virol. 2012;86:2045–2055. doi: 10.1128/JVI.06091-11. - DOI - PMC - PubMed
-
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

