The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx
- PMID: 27388563
- PMCID: PMC4936249
- DOI: 10.1186/s40168-016-0182-1
The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx
Abstract
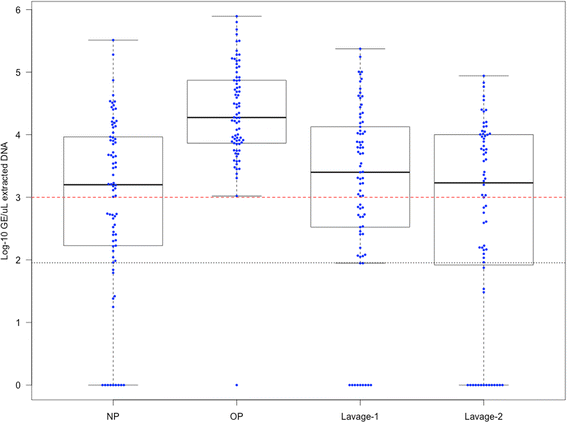
Background: Invasive methods requiring general anaesthesia are needed to sample the lung microbiota in young children who do not expectorate. This poses substantial challenges to longitudinal study of paediatric airway microbiota. Non-invasive upper airway sampling is an alternative method for monitoring airway microbiota; however, there are limited data describing the relationship of such results with lung microbiota in young children. In this study, we compared the upper and lower airway microbiota in young children to determine whether non-invasive upper airway sampling procedures provide a reliable measure of either lung microbiota or clinically defined differences.
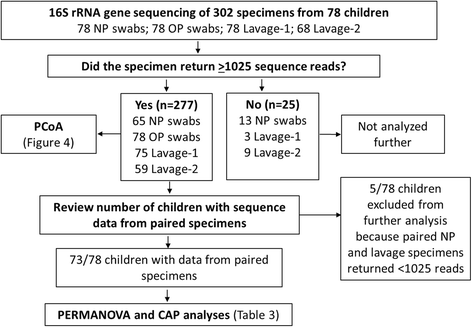
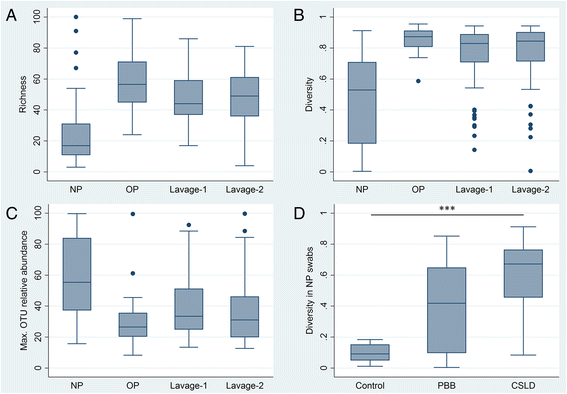
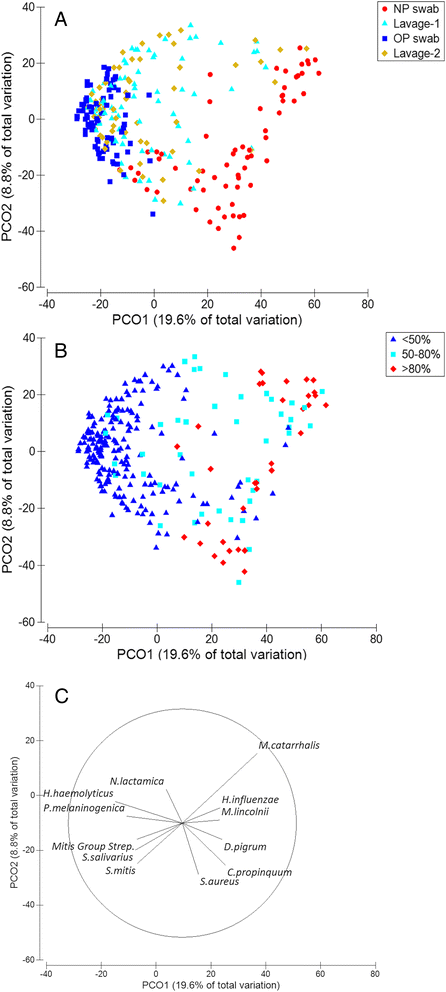
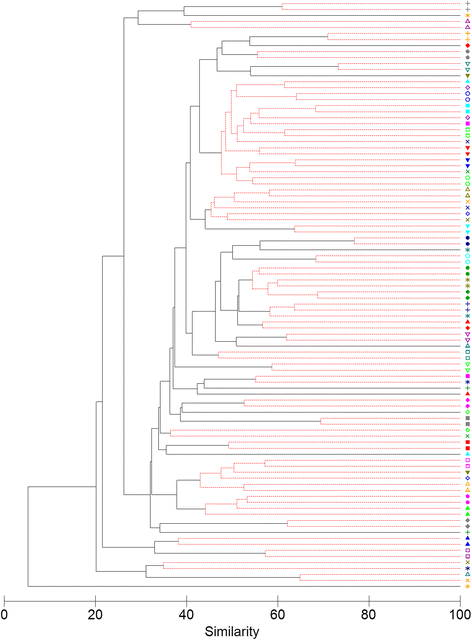
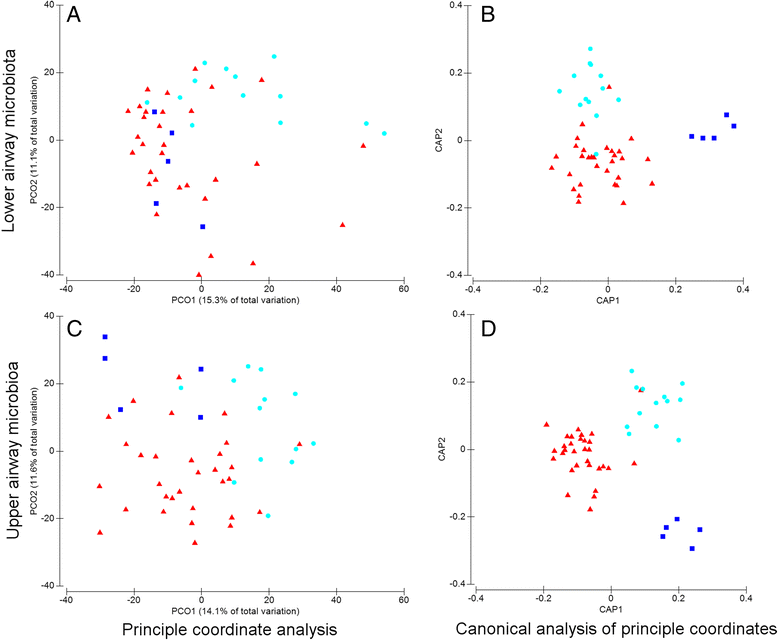
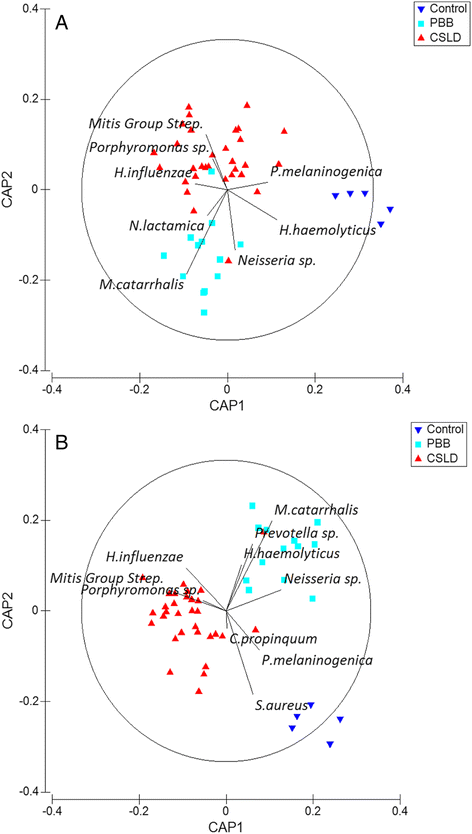
Results: The microbiota in oropharyngeal (OP) swabs, nasopharyngeal (NP) swabs and bronchoalveolar lavage (BAL) from 78 children (median age 2.2 years) with and without lung disease were characterised using 16S rRNA gene sequencing. Permutational multivariate analysis of variance (PERMANOVA) detected significant differences between the microbiota in BAL and those in both OP swabs (p = 0.0001, Pseudo-F = 12.2, df = 1) and NP swabs (p = 0.0001; Pseudo-F = 21.9, df = 1) with the NP and BAL microbiota more different than the OP and BAL, as indicated by a higher Pseudo-F value. The microbiota in combined OP and NP data (upper airways) provided a more comprehensive representation of BAL microbiota, but significant differences between the upper airway and BAL microbiota remained, albeit with a considerably smaller Pseudo-F (PERMANOVA p = 0.0001; Pseudo-F = 4.9, df = 1). Despite this overall difference, paired BAL and upper airway (OP and NP) microbiota were >50 % similar among 69 % of children. Furthermore, canonical analysis of principal coordinates (CAP analysis) detected significant differences between the microbiota from clinically defined groups when analysing either BAL (eigenvalues >0.8; misclassification rate 26.5 %) or the combined OP and NP data (eigenvalues >0.8; misclassification rate 12.2 %).
Conclusions: Upper airway sampling provided an imperfect, but reliable, representation of the BAL microbiota for most children in this study. We recommend inclusion of both OP and NP specimens when non-invasive upper airway sampling is needed to assess airway microbiota in young children who do not expectorate. The results of the CAP analysis suggest lower and upper airway microbiota profiles may differentiate children with chronic suppurative lung disease from those with persistent bacterial bronchitis; however, further research is needed to confirm this observation.
Keywords: Bronchoalveolar lavage; Lower airways; Microbiota; Nasopharynx; Oropharynx; Paediatric lung disease; Upper airways.
Figures
References
-
- Forton J. Induced sputum in young healthy children with cystic fibrosis. Paediatr Respir Rev. 2015;16(Suppl 1):6–8. - PubMed
-
- Whiteson KL, Bailey B, Bergkessel M, Conrad D, Delhaes L, Felts B, et al. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis. Parallels from island biogeography. Am J Respir Crit Care Med. 2014;189(11):1309–15. doi: 10.1164/rccm.201312-2129PP. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

