Prediction of CYP2D6 phenotype from genotype across world populations
- PMID: 27388693
- PMCID: PMC5292679
- DOI: 10.1038/gim.2016.80
Prediction of CYP2D6 phenotype from genotype across world populations
Erratum in
-
CORRIGENDUM: Prediction of CYP2D6 phenotype from genotype across world populations.Genet Med. 2016 Nov;18(11):1167. doi: 10.1038/gim.2016.160. Genet Med. 2016. PMID: 27797374 Free PMC article. No abstract available.
Abstract
Purpose: Owing to its highly polymorphic nature and major contribution to the metabolism and bioactivation of numerous clinically used drugs, CYP2D6 is one of the most extensively studied drug-metabolizing enzymes and pharmacogenes. CYP2D6 alleles confer no, decreased, normal, or increased activity and cause a wide range of activity among individuals and between populations. However, there is no standard approach to translate diplotypes into predicted phenotype.
Methods: We exploited CYP2D6 allele-frequency data that have been compiled for Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines (>60,000 subjects, 173 reports) in order to estimate genotype-predicted phenotype status across major world populations based on activity score (AS) assignments.
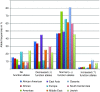
Results: Allele frequencies vary considerably across the major ethnic groups predicting poor metabolizer status (AS = 0) between 0.4 and 5.4% across world populations. The prevalence of genotypic intermediate (AS = 0.5) and normal (AS = 1, 1.5, or 2) metabolizers ranges between 0.4 and 11% and between 67 and 90%, respectively. Finally, 1 to 21% of subjects (AS >2) are predicted to have ultrarapid metabolizer status.
Conclusions: This comprehensive study summarizes allele frequencies, diplotypes, and predicted phenotype across major populations, providing a rich data resource for clinicians and researchers. Challenges of phenotype prediction from genotype data are highlighted and discussed.Genet Med 19 1, 69-76.
Figures


References
-
- Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet 2009;48:761–804. - PubMed
-
- Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet 2009;48:689–723. - PubMed
-
- He ZX, Chen XW, Zhou ZW, Zhou SF. Impact of physiological, pathological and environmental factors on the expression and activity of human cytochrome P450 2D6 and implications in precision medicine. Drug Metab Rev. 2015:1–50. - PubMed
-
- Sim SC, Ingelman-Sundberg M. Update on allele nomenclature for human cytochromes P450 and the Human Cytochrome P450 Allele (CYP-allele) Nomenclature Database. Methods Mol Biol 2013;987:251–259. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

