Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq
- PMID: 27389619
- PMCID: PMC4937404
- DOI: 10.1038/srep29345
Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq
Abstract
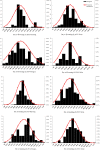
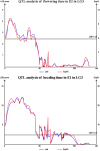
Orchardgrass (Dactylis glomerata L.) is one of the most economically important perennial, cool-season forage species grown and pastured worldwide. High-density genetic linkage mapping is a valuable and effective method for exploring complex quantitative traits. In this study, we developed 447,177 markers based on SLAF-seq and used them to perform a comparative genomics analysis. Perennial ryegrass sequences were the most similar (5.02%) to orchardgrass sequences. A high-density linkage map of orchardgrass was constructed using 2,467 SLAF markers and 43 SSRs, which were distributed on seven linkage groups spanning 715.77 cM. The average distance between adjacent markers was 0.37 cM. Based on phenotyping in four environments, 11 potentially significant quantitative trait loci (QTLs) for two target traits-heading date (HD) and flowering time (FT)-were identified and positioned on linkage groups LG1, LG3, and LG5. Significant QTLs explained 8.20-27.00% of the total phenotypic variation, with the LOD ranging from 3.85-12.21. Marker167780 and Marker139469 were associated with FT and HD at the same location (Ya'an) over two different years. The utility of SLAF markers for rapid generation of genetic maps and QTL analysis has been demonstrated for heading date and flowering time in a global forage grass.
Figures

Similar articles
-
A genetic linkage map of tetraploid orchardgrass (Dactylis glomerata L.) and quantitative trait loci for heading date.Genome. 2012 May;55(5):360-9. doi: 10.1139/g2012-026. Epub 2012 May 3. Genome. 2012. PMID: 22551303
-
High-density genetic map construction and QTLs identification for plant height in white jute (Corchorus capsularis L.) using specific locus amplified fragment (SLAF) sequencing.BMC Genomics. 2017 May 8;18(1):355. doi: 10.1186/s12864-017-3712-8. BMC Genomics. 2017. PMID: 28482802 Free PMC article.
-
Construction of the first high-density genetic linkage map and identification of seed yield-related QTLs and candidate genes in Elymus sibiricus, an important forage grass in Qinghai-Tibet Plateau.BMC Genomics. 2019 Nov 14;20(1):861. doi: 10.1186/s12864-019-6254-4. BMC Genomics. 2019. PMID: 31726988 Free PMC article.
-
Genetic maps of SSR and SRAP markers in diploid orchardgrass (Dactylis glomerata L.) using the pseudo-testcross strategy.Genome. 2011 Mar;54(3):212-21. doi: 10.1139/G10-111. Genome. 2011. PMID: 21423284
-
Development of a high-density genetic linkage map and identification of flowering time QTLs in adzuki bean (Vigna angularis).Sci Rep. 2016 Dec 23;6:39523. doi: 10.1038/srep39523. Sci Rep. 2016. PMID: 28008173 Free PMC article.
Cited by
-
Genome assembly provides insights into the genome evolution and flowering regulation of orchardgrass.Plant Biotechnol J. 2020 Feb;18(2):373-388. doi: 10.1111/pbi.13205. Epub 2019 Jul 30. Plant Biotechnol J. 2020. PMID: 31276273 Free PMC article.
-
High-Density Genetic Map Construction and Identification of QTLs Controlling Oleic and Linoleic Acid in Peanut using SLAF-seq and SSRs.Sci Rep. 2018 Apr 3;8(1):5479. doi: 10.1038/s41598-018-23873-7. Sci Rep. 2018. PMID: 29615772 Free PMC article.
-
Genome-wide association study identifying genetic variants associated with carcass backfat thickness, lean percentage and fat percentage in a four-way crossbred pig population using SLAF-seq technology.BMC Genomics. 2022 Aug 15;23(1):594. doi: 10.1186/s12864-022-08827-8. BMC Genomics. 2022. PMID: 35971078 Free PMC article.
-
QTL mapping for growth-related traits by constructing the first genetic linkage map in Simao pine.BMC Plant Biol. 2022 Jan 22;22(1):48. doi: 10.1186/s12870-022-03425-y. BMC Plant Biol. 2022. PMID: 35065611 Free PMC article.
-
Construction of a high-density genetic map based on large-scale marker development in Coix lacryma-jobi L. using specific-locus amplified fragment sequencing (slaf-seq).Sci Rep. 2024 Apr 26;14(1):9606. doi: 10.1038/s41598-024-58167-8. Sci Rep. 2024. PMID: 38670987 Free PMC article.
References
-
- Stewart A. & Ellison N. Dactylis. In Wild Crop Relatives: Genomic and Breeding Resources, Springer Berlin Heidelberg, 73–87 (2011).
-
- Hirata M., Yuyama N. & Cai H. Isolation and characterization of simple sequence repeat markers for the tetraploid forage grass Dactylis glomerata. Plant breeding 130, 503–506 (2011).
-
- Hannaway D. et al.. Orchardgrass (dactylis glomerata L.). ([Corvallis, Or.]: Oregon State University Extension Service;[Olympia, Wash.]: Washington State University Cooperative Extension;[Moscow, Idaho]: University of Idaho Cooperative Extension System;[Washington, DC]: US Dept. of Agriculture, 1999).
-
- Bushman B. S., Robins J. G. & Jensen K. B. Dry matter yield, heading date, and plant mortality of orchardgrass subspecies in a semiarid environment. Crop Science 52, 745–751 (2012).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

