Separate F-Type Plasmids Have Shaped the Evolution of the H30 Subclone of Escherichia coli Sequence Type 131
- PMID: 27390780
- PMCID: PMC4933990
- DOI: 10.1128/mSphere.00121-16
Separate F-Type Plasmids Have Shaped the Evolution of the H30 Subclone of Escherichia coli Sequence Type 131
Abstract
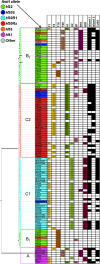
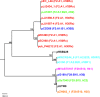
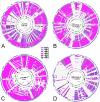
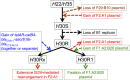
The extraintestinal pathogenic Escherichia coli (ExPEC) H30 subclone of sequence type 131 (ST131-H30) has emerged abruptly as a dominant lineage of ExPEC responsible for human disease. The ST131-H30 lineage has been well described phylogenetically, yet its plasmid complement is not fully understood. Here, single-molecule, real-time sequencing was used to generate the complete plasmid sequences of ST131-H30 isolates and those belonging to other ST131 clades. Comparative analyses revealed separate F-type plasmids that have shaped the evolution of the main fluoroquinolone-resistant ST131-H30 clades. Specifically, an F1:A2:B20 plasmid is strongly associated with the H30R/C1 clade, whereas an F2:A1:B- plasmid is associated with the H30Rx/C2 clade. A series of plasmid gene losses, gains, and rearrangements involving IS26 likely led to the current plasmid complements within each ST131-H30 sublineage, which contain several overlapping gene clusters with putative functions in virulence and fitness, suggesting plasmid-mediated convergent evolution. Evidence suggests that the H30Rx/C2-associated F2:A1:B- plasmid type was present in strains ancestral to the acquisition of fluoroquinolone resistance and prior to the introduction of a multidrug resistance-encoding gene cassette harboring bla CTX-M-15. In vitro experiments indicated a host strain-independent low frequency of plasmid transfer, differential levels of plasmid stability even between closely related ST131-H30 strains, and possible epistasis for carriage of these plasmids within the H30R/Rx lineages. IMPORTANCE A clonal lineage of Escherichia coli known as ST131 has emerged as a dominating strain type causing extraintestinal infections in humans. The evolutionary history of ST131 E. coli is now well understood. However, the role of plasmids in ST131's evolutionary history is poorly defined. This study utilized real-time, single-molecule sequencing to compare plasmids from various current and historical lineages of ST131. From this work, it was determined that a series of plasmid gains, losses, and recombinational events has led to the currently circulating plasmids of ST131 strains. These plasmids appear to have evolved to acquire similar gene clusters on multiple occasions, suggesting possible plasmid-mediated convergent evolution leading to evolutionary success. These plasmids also appear to be better suited to exist in specific strains of ST131 due to coadaptive mutations. Overall, a series of events has enabled the evolution of ST131 plasmids, possibly contributing to the lineage's success.
Keywords: Escherichia coli; ST131; genomes; plasmids.
Figures



References
-
- Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi:10.1093/infdis/jis933. - DOI - PMC - PubMed
-
- Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi:10.1128/mBio.00377-13. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
