Characterization of a novel zebrafish (Danio rerio) gene, wdr81, associated with cerebellar ataxia, mental retardation and dysequilibrium syndrome (CAMRQ)
- PMID: 27390838
- PMCID: PMC4690267
- DOI: 10.1186/s12868-015-0229-4
Characterization of a novel zebrafish (Danio rerio) gene, wdr81, associated with cerebellar ataxia, mental retardation and dysequilibrium syndrome (CAMRQ)
Abstract
Background: WDR81 (WD repeat-containing protein 81) is associated with cerebellar ataxia, mental retardation and disequilibrium syndrome (CAMRQ2, [MIM 610185]). Human and mouse studies suggest that it might be a gene of importance during neurodevelopment. This study aimed at fully characterizing the structure of the wdr81 transcript, detecting the possible transcript variants and revealing its expression profile in zebrafish, a powerful model organism for studying development and disease.
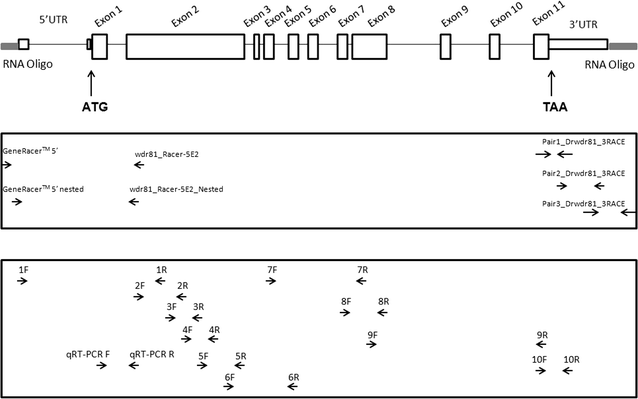
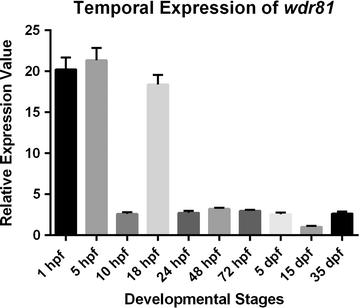
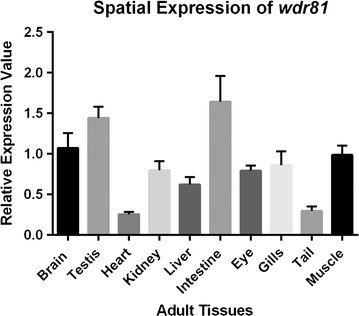
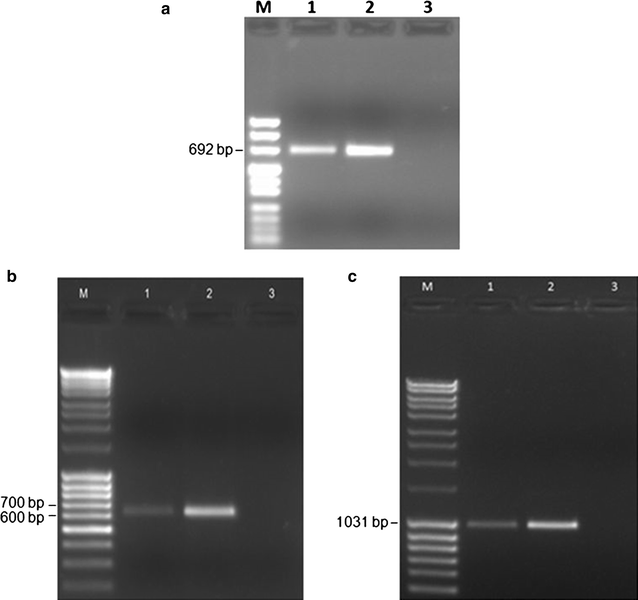
Results: As expected in human and mouse orthologous proteins, zebrafish wdr81 is predicted to possess a BEACH (Beige and Chediak-Higashi) domain, a major facilitator superfamily domain and WD40-repeats, which indicates a conserved function in these species. We observed that zebrafish wdr81 encodes one open reading frame while the transcript has one 5' untranslated region (UTR) and the prediction of the 3' UTR was mainly confirmed along with a detected insertion site in the embryo and adult brain. This insertion site was also found in testis, heart, liver, eye, tail and muscle, however, there was no amplicon in kidney, intestine and gills, which might be the result of possible alternative polyadenylation processes among tissues. The 5 and 18 hpf were critical timepoints of development regarding wdr81 expression. Furthermore, the signal of the RNA probe was stronger in the eye and brain at 18 and 48 hpf, then decreased at 72 hpf. Finally, expression of wdr81 was detected in the adult brain and eye tissues, including but not restricted to photoreceptors of the retina, presumptive Purkinje cells and some neurogenic brains regions.
Conclusions: Taken together these data emphasize the importance of this gene during neurodevelopment and a possible role for neuronal proliferation. Our data provide a basis for further studies to fully understand the function of wdr81.
Keywords: In situ hybridization; RACE; Zebrafish; qRT-PCR; wdr81.
Figures





References
-
- Gulsuner S, Tekinay AB, Doerschner K, Boyaci H, Bilguvar K, Unal H, Onat OE, Atalar E, Basak N, Topaloglu H, Kansu T, Tan M, Tan U, Gunel M, Ozcelik T. Homozygosity mapping and targeted genomic sequencing reveal the gene responsible for cerebellar hypoplasia and quadrupedal locomotion in a consanguineous kindred. Genome Res. 2011;21(12):1995–2003. doi: 10.1101/gr.126110.111. - DOI - PMC - PubMed
-
- Donnard E, Asprino PF, Correa BR, Bettoni F, Koyama FC, Navarro FCP, Perez RO, Mariadason J, Sieber OM, Strausberg RL, Simpson AJG, Jardim DLF, Reis LFL, Parmigiani RB, Galante PAF, Camargo AA. Mutational analysis of genes coding for cell surface proteins in colorectal cancer cell lines reveal novel altered pathways, druggable mutations and mutated epitopes for targeted therapy. Oncotarget. 2014;5(19):9199–9213. doi: 10.18632/oncotarget.2374. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

