Mechanisms of ATP-Dependent Chromatin Remodeling Motors
- PMID: 27391925
- PMCID: PMC9157391
- DOI: 10.1146/annurev-biophys-051013-022819
Mechanisms of ATP-Dependent Chromatin Remodeling Motors
Abstract
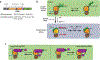
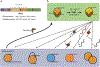
Chromatin remodeling motors play essential roles in all DNA-based processes. These motors catalyze diverse outcomes ranging from sliding the smallest units of chromatin, known as nucleosomes, to completely disassembling chromatin. The broad range of actions carried out by these motors on the complex template presented by chromatin raises many stimulating mechanistic questions. Other well-studied nucleic acid motors provide examples of the depth of mechanistic understanding that is achievable from detailed biophysical studies. We use these studies as a guiding framework to discuss the current state of knowledge of chromatin remodeling mechanisms and highlight exciting open questions that would continue to benefit from biophysical analyses.
Keywords: ATPase; INO80; ISWI; SWI/SNF; SWR; histones; molecular motors; nucleosome.
Figures




References
-
- Aalfs JD, Narlikar GJ, Kingston RE. 2001. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem 276(36):34270–78 - PubMed
-
- Adamkewicz JI, Mueller CG, Hansen KE, Prud’homme WA, Thorner J. 2000. Purification and enzymic properties of Mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J. Biol. Chem 275(28):21158–68 - PubMed
-
- Anderson JD, Widom J. 2000. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol 296(4):979–87 - PubMed
-
- Andrews AJ, Luger K. 2011. Nucleosome structure(s) and stability: variations on a theme. Annu. Rev. Biophys 40:99–117 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

