Nuclear Noncoding RNAs and Genome Stability
- PMID: 27392145
- PMCID: PMC5136474
- DOI: 10.1016/j.molcel.2016.06.011
Nuclear Noncoding RNAs and Genome Stability
Abstract
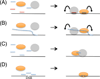
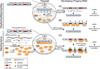
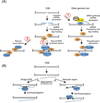
In modern molecular biology, RNA has emerged as a versatile macromolecule capable of mediating an astonishing number of biological functions beyond its role as a transient messenger of genetic information. The recent discovery and functional analyses of new classes of noncoding RNAs (ncRNAs) have revealed their widespread use in many pathways, including several in the nucleus. This Review focuses on the mechanisms by which nuclear ncRNAs directly contribute to the maintenance of genome stability. We discuss how ncRNAs inhibit spurious recombination among repetitive DNA elements, repress mobilization of transposable elements (TEs), template or bridge DNA double-strand breaks (DSBs) during repair, and direct developmentally regulated genome rearrangements in some ciliates. These studies reveal an unexpected repertoire of mechanisms by which ncRNAs contribute to genome stability and even potentially fuel evolution by acting as templates for genome modification.
Keywords: DNA double-strand break (DSB); DNA repair; diRNA; genome stability; heterochromatin; lncRNA; noncoding RNA (ncRNA); piRNA; siRNA; transposon silencing.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

References
-
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular cell. 2012;46:115–124. - PubMed
-
- Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics. 2013;12:254–278. - PubMed
-
- Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nature structural & molecular biology. 2012;19:948–956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

