Origins, structures, and functions of circulating DNA in oncology
- PMID: 27392603
- PMCID: PMC5035665
- DOI: 10.1007/s10555-016-9629-x
Origins, structures, and functions of circulating DNA in oncology
Abstract
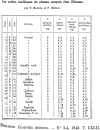
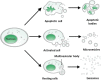
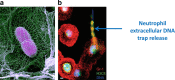
While various clinical applications especially in oncology are now in progress such as diagnosis, prognosis, therapy monitoring, or patient follow-up, the determination of structural characteristics of cell-free circulating DNA (cirDNA) are still being researched. Nevertheless, some specific structures have been identified and cirDNA has been shown to be composed of many "kinds." This structural description goes hand-in-hand with the mechanisms of its origins such as apoptosis, necrosis, active release, phagocytosis, and exocytose. There are multiple structural forms of cirDNA depending upon the mechanism of release: particulate structures (exosomes, microparticles, apoptotic bodies) or macromolecular structures (nucleosomes, virtosomes/proteolipidonucleic acid complexes, DNA traps, links with serum proteins or to the cell-free membrane parts). In addition, cirDNA concerns both nuclear and/or mitochondrial DNA with both species exhibiting different structural characteristics that potentially reveal different forms of biological stability or diagnostic significance. This review focuses on the origins, structures and functional aspects that are paradoxically less well described in the literature while numerous reviews are directed to the clinical application of cirDNA. Differentiation of the various structures and better knowledge of the fate of cirDNA would considerably expand the diagnostic power of cirDNA analysis especially with regard to the patient follow-up enlarging the scope of personalized medicine. A better understanding of the subsequent fate of cirDNA would also help in deciphering its functional aspects such as their capacity for either genometastasis or their pro-inflammatory and immunological effects.
Keywords: Cancer; Cell-free circulating DNA; Functions; Origins; Structures.
Conflict of interest statement
The authors declare that they have are no conflicts of interest.
Figures

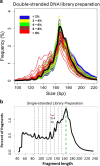
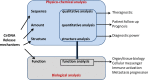
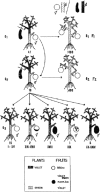
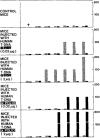




References
-
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochimica et Biophysica Acta. 2007;1775(1):181–232. - PubMed
-
- Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum. Recent developments. Annals of the New York Academy of Sciences. 2008;1137:1–6. - PubMed
-
- Miescher F. Ueber die chemische Zusammensetzung der Eiterzellen. Medicinisch-chemische Untersuchungen. 1871;4:441–460.
-
- Levene PA, Haller HL. On the configurational relationship of 3-Chlorobutyric and 3-Hydroxybutyric acids. Science (New York, N.Y.) 1929;69(1776):47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

