Mechanisms of viral mutation
- PMID: 27392606
- PMCID: PMC5075021
- DOI: 10.1007/s00018-016-2299-6
Mechanisms of viral mutation
Abstract
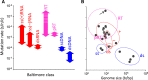
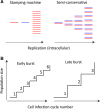
The remarkable capacity of some viruses to adapt to new hosts and environments is highly dependent on their ability to generate de novo diversity in a short period of time. Rates of spontaneous mutation vary amply among viruses. RNA viruses mutate faster than DNA viruses, single-stranded viruses mutate faster than double-strand virus, and genome size appears to correlate negatively with mutation rate. Viral mutation rates are modulated at different levels, including polymerase fidelity, sequence context, template secondary structure, cellular microenvironment, replication mechanisms, proofreading, and access to post-replicative repair. Additionally, massive numbers of mutations can be introduced by some virus-encoded diversity-generating elements, as well as by host-encoded cytidine/adenine deaminases. Our current knowledge of viral mutation rates indicates that viral genetic diversity is determined by multiple virus- and host-dependent processes, and that viral mutation rates can evolve in response to specific selective pressures.
Keywords: Evolution; Genetic diversity; Hyper-mutation; Mutation rate; Post-replicative repair; Replication fidelity; Virus.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

