Transcriptome Analysis of Capsicum Chlorosis Virus-Induced Hypersensitive Resistance Response in Bell Capsicum
- PMID: 27398596
- PMCID: PMC4939944
- DOI: 10.1371/journal.pone.0159085
Transcriptome Analysis of Capsicum Chlorosis Virus-Induced Hypersensitive Resistance Response in Bell Capsicum
Abstract
Background: Capsicum chlorosis virus (CaCV) is an emerging pathogen of capsicum, tomato and peanut crops in Australia and South-East Asia. Commercial capsicum cultivars with CaCV resistance are not yet available, but CaCV resistance identified in Capsicum chinense is being introgressed into commercial Bell capsicum. However, our knowledge of the molecular mechanisms leading to the resistance response to CaCV infection is limited. Therefore, transcriptome and expression profiling data provide an important resource to better understand CaCV resistance mechanisms.
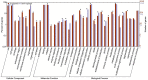
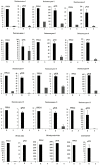
Methodology/principal findings: We assembled capsicum transcriptomes and analysed gene expression using Illumina HiSeq platform combined with a tag-based digital gene expression system. Total RNA extracted from CaCV/mock inoculated CaCV resistant (R) and susceptible (S) capsicum at the time point when R line showed a strong hypersensitive response to CaCV infection was used in transcriptome assembly. Gene expression profiles of R and S capsicum in CaCV- and buffer-inoculated conditions were compared. None of the genes were differentially expressed (DE) between R and S cultivars when mock-inoculated, while 2484 genes were DE when inoculated with CaCV. Functional classification revealed that the most highly up-regulated DE genes in R capsicum included pathogenesis-related genes, cell death-associated genes, genes associated with hormone-mediated signalling pathways and genes encoding enzymes involved in synthesis of defense-related secondary metabolites. We selected 15 genes to confirm DE expression levels by real-time quantitative PCR.
Conclusion/significance: DE transcript profiling data provided comprehensive gene expression information to gain an understanding of the underlying CaCV resistance mechanisms. Further, we identified candidate CaCV resistance genes in the CaCV-resistant C. annuum x C. chinense breeding line. This knowledge will be useful in future for fine mapping of the CaCV resistance locus and potential genetic engineering of resistance into CaCV-susceptible crops.
Conflict of interest statement
Figures

Similar articles
-
Effects of Elevated Temperature on the Susceptibility of Capsicum Plants to Capsicum Chlorosis Virus Infection.Pathogens. 2022 Feb 2;11(2):200. doi: 10.3390/pathogens11020200. Pathogens. 2022. PMID: 35215143 Free PMC article.
-
Involvement of MicroRNAs in the Hypersensitive Response of Capsicum Plants to the Capsicum Chlorosis Virus at Elevated Temperatures.Pathogens. 2024 Aug 31;13(9):745. doi: 10.3390/pathogens13090745. Pathogens. 2024. PMID: 39338939 Free PMC article.
-
Intracellular Localization, Interactions and Functions of Capsicum Chlorosis Virus Proteins.Front Microbiol. 2017 Apr 11;8:612. doi: 10.3389/fmicb.2017.00612. eCollection 2017. Front Microbiol. 2017. PMID: 28443083 Free PMC article.
-
Functional roles of the pepper leucine-rich repeat protein and its interactions with pathogenesis-related and hypersensitive-induced proteins in plant cell death and immunity.Planta. 2017 Sep;246(3):351-364. doi: 10.1007/s00425-017-2709-5. Epub 2017 May 15. Planta. 2017. PMID: 28508261 Review.
-
Spicing Up the N Gene: F. O. Holmes and Tobacco mosaic virus Resistance in Capsicum and Nicotiana Plants.Phytopathology. 2017 Feb;107(2):148-157. doi: 10.1094/PHYTO-07-16-0264-RVW. Epub 2016 Nov 30. Phytopathology. 2017. PMID: 27642796 Review.
Cited by
-
Glutathione Can Compensate for Salicylic Acid Deficiency in Tobacco to Maintain Resistance to Tobacco Mosaic Virus.Front Plant Sci. 2019 Sep 13;10:1115. doi: 10.3389/fpls.2019.01115. eCollection 2019. Front Plant Sci. 2019. PMID: 31608082 Free PMC article.
-
Overview of Biotic Stresses in Pepper (Capsicum spp.): Sources of Genetic Resistance, Molecular Breeding and Genomics.Int J Mol Sci. 2020 Apr 8;21(7):2587. doi: 10.3390/ijms21072587. Int J Mol Sci. 2020. PMID: 32276403 Free PMC article. Review.
-
Pest categorisation of Capsicum chlorosis virus.EFSA J. 2022 Jun 14;20(6):e07337. doi: 10.2903/j.efsa.2022.7337. eCollection 2022 Jun. EFSA J. 2022. PMID: 35734283 Free PMC article.
-
Effects of Elevated Temperature on the Susceptibility of Capsicum Plants to Capsicum Chlorosis Virus Infection.Pathogens. 2022 Feb 2;11(2):200. doi: 10.3390/pathogens11020200. Pathogens. 2022. PMID: 35215143 Free PMC article.
-
The Defense Response of Nicotiana benthamiana to Peanut Stunt Virus Infection in the Presence of Symptom Exacerbating Satellite RNA.Viruses. 2018 Aug 23;10(9):449. doi: 10.3390/v10090449. Viruses. 2018. PMID: 30142955 Free PMC article.
References
-
- Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, et al., editors. The evolution of chili peppers (Capsicum-Solanaceae): a cytogenetic perspective. VI International Solanaceae Conference: Genomics Meets Biodiversity 745; 2006.
-
- Persley D, Cooke T, House S. Diseases of vegetable crops in Australia: CSIRO PUBLISHING; 2010.
-
- Reddy M, Srivastava A, Kumar S, Kumar R, Chawda N, Ebert A, et al. Chilli (Capsicum annuum L.) breeding in India: an overview. SABRAO Journal of Breeding and Genetics. 2014;46(2):160–73.
-
- Persley D, Thomas J, Sharman M. Tospoviruses—an Australian perspective. Australasian Plant Pathology. 2006;35(2):161–80.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

