Lipids modulate the conformational dynamics of a secondary multidrug transporter
- PMID: 27399258
- PMCID: PMC5248563
- DOI: 10.1038/nsmb.3262
Lipids modulate the conformational dynamics of a secondary multidrug transporter
Abstract
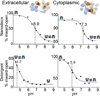
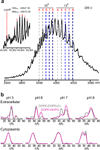
Direct interactions with lipids have emerged as key determinants of the folding, structure and function of membrane proteins, but an understanding of how lipids modulate protein dynamics is still lacking. Here, we systematically explored the effects of lipids on the conformational dynamics of the proton-powered multidrug transporter LmrP from Lactococcus lactis, using the pattern of distances between spin-label pairs previously shown to report on alternating access of the protein. We uncovered, at the molecular level, how the lipid headgroups shape the conformational-energy landscape of the transporter. The model emerging from our data suggests a direct interaction between lipid headgroups and a conserved motif of charged residues that control the conformational equilibrium through an interplay of electrostatic interactions within the protein. Together, our data lay the foundation for a comprehensive model of secondary multidrug transport in lipid bilayers.
Figures




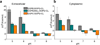
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

