EPHB4 kinase-inactivating mutations cause autosomal dominant lymphatic-related hydrops fetalis
- PMID: 27400125
- PMCID: PMC4966301
- DOI: 10.1172/JCI85794
EPHB4 kinase-inactivating mutations cause autosomal dominant lymphatic-related hydrops fetalis
Abstract
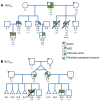
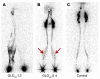
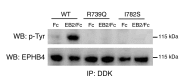
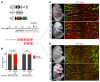
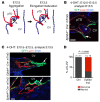
Hydrops fetalis describes fluid accumulation in at least 2 fetal compartments, including abdominal cavities, pleura, and pericardium, or in body tissue. The majority of hydrops fetalis cases are nonimmune conditions that present with generalized edema of the fetus, and approximately 15% of these nonimmune cases result from a lymphatic abnormality. Here, we have identified an autosomal dominant, inherited form of lymphatic-related (nonimmune) hydrops fetalis (LRHF). Independent exome sequencing projects on 2 families with a history of in utero and neonatal deaths associated with nonimmune hydrops fetalis uncovered 2 heterozygous missense variants in the gene encoding Eph receptor B4 (EPHB4). Biochemical analysis determined that the mutant EPHB4 proteins are devoid of tyrosine kinase activity, indicating that loss of EPHB4 signaling contributes to LRHF pathogenesis. Further, inactivation of Ephb4 in lymphatic endothelial cells of developing mouse embryos led to defective lymphovenous valve formation and consequent subcutaneous edema. Together, these findings identify EPHB4 as a critical regulator of early lymphatic vascular development and demonstrate that mutations in the gene can cause an autosomal dominant form of LRHF that is associated with a high mortality rate.
Figures

References
-
- Bellini C, et al. Etiology of non-immune hydrops fetalis: an update. Am J Med Genet A. 2015;167A(5):1082–1088. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

