Stochastic alternative splicing is prevalent in mungbean (Vigna radiata)
- PMID: 27400146
- PMCID: PMC5258860
- DOI: 10.1111/pbi.12600
Stochastic alternative splicing is prevalent in mungbean (Vigna radiata)
Abstract
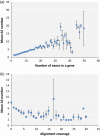
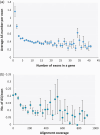
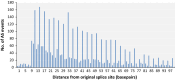
Alternative splicing (AS) can produce multiple mature mRNAs from the same primary transcript, thereby generating diverse proteins and phenotypes from the same gene. To assess the prevalence of AS in mungbean (Vigna radiata), we analysed whole-genome RNA sequencing data from root, leaf, flower and pod tissues and found that at least 37.9% of mungbean genes are subjected to AS. The number of AS transcripts exhibited a strong correlation with exon number and thus resembled a uniform probabilistic event rather than a specific regulatory function. The proportion of frameshift splicing was close to the expected frequency of random splicing. However, alternative donor and acceptor AS events tended to occur at multiples of three nucleotides (i.e. the codon length) from the main splice site. Genes with high exon number and expression level, which should have the most AS if splicing is purely stochastic, exhibited less AS, implying the existence of negative selection against excessive random AS. Functional AS is probably rare: a large proportion of AS isoforms exist at very low copy per cell on average or are expressed at much lower levels than default transcripts. Conserved AS was only detected in 629 genes (2.8% of all genes in the genome) when compared to Vigna angularis, and in 16 genes in more distant species like soya bean. These observations highlight the challenges of finding and cataloguing candidates for experimentally proven AS isoforms in a crop genome.
Keywords: RNA sequencing; alternative splicing; evolutionary conservation; mungbean (Vigna radiata); stochastic process.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

Similar articles
-
The complete mitochondrial genome of mungbean Vigna radiata var. radiata NM92 and a phylogenetic analysis of crops in angiosperms.Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Sep;27(5):3731-2. doi: 10.3109/19401736.2015.1079879. Epub 2015 Oct 15. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26469726
-
A chromosome-scale assembly of the black gram (Vigna mungo) genome.Mol Ecol Resour. 2021 Jan;21(1):238-250. doi: 10.1111/1755-0998.13243. Epub 2020 Sep 8. Mol Ecol Resour. 2021. PMID: 32794377
-
Construction of genetic linkage map and genome dissection of domestication-related traits of moth bean (Vigna aconitifolia), a legume crop of arid areas.Mol Genet Genomics. 2019 Jun;294(3):621-635. doi: 10.1007/s00438-019-01536-0. Epub 2019 Feb 9. Mol Genet Genomics. 2019. PMID: 30739203
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
-
Multiplexed primer extension sequencing: A targeted RNA-seq method that enables high-precision quantitation of mRNA splicing isoforms and rare pre-mRNA splicing intermediates.Methods. 2020 Apr 1;176:34-45. doi: 10.1016/j.ymeth.2019.05.013. Epub 2019 May 21. Methods. 2020. PMID: 31121301 Free PMC article. Review.
Cited by
-
Genome-wide differences in gene expression and alternative splicing in developing embryo and endosperm, and between F1 hybrids and their parental pure lines in sorghum.Plant Mol Biol. 2022 Jan;108(1-2):1-14. doi: 10.1007/s11103-021-01196-y. Epub 2021 Nov 30. Plant Mol Biol. 2022. PMID: 34846608
-
Evolution of Alternative Splicing in Eudicots.Front Plant Sci. 2019 Jun 12;10:707. doi: 10.3389/fpls.2019.00707. eCollection 2019. Front Plant Sci. 2019. PMID: 31244865 Free PMC article.
-
Unveiling the Complexity of Red Clover (Trifolium pratense L.) Transcriptome and Transcriptional Regulation of Isoflavonoid Biosynthesis Using Integrated Long- and Short-Read RNAseq.Int J Mol Sci. 2021 Nov 23;22(23):12625. doi: 10.3390/ijms222312625. Int J Mol Sci. 2021. PMID: 34884432 Free PMC article.
-
Alternative splicing shapes the transcriptome complexity in blackgram [Vigna mungo (L.) Hepper].Funct Integr Genomics. 2023 May 3;23(2):144. doi: 10.1007/s10142-023-01066-4. Funct Integr Genomics. 2023. PMID: 37133618
-
Integrative expression network analysis of microRNA and gene isoforms in sacred lotus.BMC Genomics. 2020 Jun 25;21(1):429. doi: 10.1186/s12864-020-06853-y. BMC Genomics. 2020. PMID: 32586276 Free PMC article.
References
-
- Barbazuk, W.B. , Fu, Y. and McGinnis, K.M. (2008) Genome‐wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18, 1381–1392. - PubMed
-
- Breitbart, R.E. , Andreadis, A. and Nadal‐Ginard, B. (1987) Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 56, 467–495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

