Tumor-induced stromal reprogramming drives lymph node transformation
- PMID: 27400148
- PMCID: PMC4994871
- DOI: 10.1038/ni.3492
Tumor-induced stromal reprogramming drives lymph node transformation
Abstract
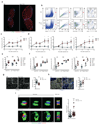
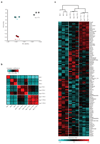
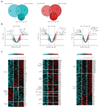
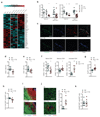
Lymph node (LN) stromal cells, particularly fibroblastic reticular cells (FRCs), provide critical structural support and regulate immunity, tolerance and the transport properties of LNs. For many tumors, metastasis to the LNs is predictive of poor prognosis. However, the stromal contribution to the evolving microenvironment of tumor-draining LNs (TDLNs) remains poorly understood. Here we found that FRCs specifically of TDLNs proliferated in response to tumor-derived cues and that the network they formed was remodeled. Comparative transcriptional analysis of FRCs from non-draining LNs and TDLNs demonstrated reprogramming of key pathways, including matrix remodeling, chemokine and/or cytokine signaling, and immunological functions such as the recruitment, migration and activation of leukocytes. In particular, downregulation of the expression of FRC-derived chemokine CCL21 and cytokine IL-7 were accompanied by altered composition and aberrant localization of immune-cell populations. Our data indicate that following exposure to tumor-derived factors, the stroma of TDLNs adapts on multiple levels to exhibit features typically associated with immunosuppression.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Comment in
-
Lymph node stroma join the cancer support network.Cell Death Differ. 2016 Dec;23(12):1899-1901. doi: 10.1038/cdd.2016.103. Epub 2016 Sep 30. Cell Death Differ. 2016. PMID: 27689877 Free PMC article. No abstract available.
Similar articles
-
Single-cell analysis reveals immune modulation and metabolic switch in tumor-draining lymph nodes.Oncoimmunology. 2020 Oct 19;9(1):1830513. doi: 10.1080/2162402X.2020.1830513. Oncoimmunology. 2020. PMID: 33117603 Free PMC article.
-
Tumor-Derived Lactic Acid Modulates Activation and Metabolic Status of Draining Lymph Node Stroma.Cancer Immunol Res. 2022 Apr 1;10(4):482-497. doi: 10.1158/2326-6066.CIR-21-0778. Cancer Immunol Res. 2022. PMID: 35362044 Free PMC article.
-
Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21.Science. 2010 May 7;328(5979):749-52. doi: 10.1126/science.1185837. Epub 2010 Mar 25. Science. 2010. PMID: 20339029
-
Role of lymph node stroma and microenvironment in T cell tolerance.Immunol Rev. 2019 Nov;292(1):9-23. doi: 10.1111/imr.12799. Epub 2019 Sep 19. Immunol Rev. 2019. PMID: 31538349 Free PMC article. Review.
-
Lymph node fibroblast-produced extracellular matrix shapes immune function.Trends Immunol. 2025 Mar;46(3):229-243. doi: 10.1016/j.it.2025.02.002. Epub 2025 Feb 28. Trends Immunol. 2025. PMID: 40023738 Review.
Cited by
-
Targeting cancer homing into the lymph node with a novel anti-CCR7 therapeutic antibody: the paradigm of CLL.MAbs. 2021 Jan-Dec;13(1):1917484. doi: 10.1080/19420862.2021.1917484. MAbs. 2021. PMID: 33944659 Free PMC article.
-
Immunotherapy: breaching the barriers for cancer treatment.Philos Trans R Soc Lond B Biol Sci. 2019 Aug 19;374(1779):20180214. doi: 10.1098/rstb.2018.0214. Epub 2019 Jul 1. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31431180 Free PMC article. Review.
-
Same yet different - how lymph node heterogeneity affects immune responses.Nat Rev Immunol. 2024 May;24(5):358-374. doi: 10.1038/s41577-023-00965-8. Epub 2023 Dec 14. Nat Rev Immunol. 2024. PMID: 38097778 Review.
-
Lymph node stroma join the cancer support network.Cell Death Differ. 2016 Dec;23(12):1899-1901. doi: 10.1038/cdd.2016.103. Epub 2016 Sep 30. Cell Death Differ. 2016. PMID: 27689877 Free PMC article. No abstract available.
-
B Cells and IL-21-Producing Follicular Helper T Cells Cooperate to Determine the Dynamic Alterations of Premetastatic Tumor Draining Lymph Nodes of Breast Cancer.Research (Wash D C). 2024 Mar 29;7:0346. doi: 10.34133/research.0346. eCollection 2024. Research (Wash D C). 2024. PMID: 38559676 Free PMC article.
References
-
- Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nature immunology. 2007;8:1255–1265. - PubMed
-
- Schumann K, et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity. 2010;32:703–713. - PubMed
-
- Roozendaal R, Mebius RE, Kraal G. The conduit system of the lymph node. International immunology. 2008;20:1483–1487. - PubMed
-
- Gretz JE, Norbury CC, Anderson AO, Proudfoot AE, Shaw S. Lymph-borne chemokines and other low molecular weight molecules reach high endothelial venules via specialized conduits while a functional barrier limits access to the lymphocyte microenvironments in lymph node cortex. J Exp Med. 2000;192:1425–1440. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

