Crystal structure and analysis of HdaB: The enteroaggregative Escherichia coli AAF/IV pilus tip protein
- PMID: 27400770
- PMCID: PMC5029526
- DOI: 10.1002/pro.2982
Crystal structure and analysis of HdaB: The enteroaggregative Escherichia coli AAF/IV pilus tip protein
Abstract
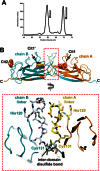
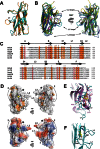
Enteroaggregative Escherichia coli is the primary cause of pediatric diarrhea in developing countries. They utilize aggregative adherence fimbriae (AAFs) to promote initial adherence to the host intestinal mucosa, promote the formation of biofilms, and mediate host invasion. Five AAFs have been identified to date and AAF/IV is amongst the most prevalent found in clinical isolates. Here we present the X-ray crystal structure of the AAF/IV tip protein HdaB at 2.0 Å resolution. It shares high structural homology with members of the Afa/Dr superfamily of fimbriae, which are involved in host invasion. We highlight surface exposed residues that share sequence homology and propose that these may function in invasion and also non-conserved regions that could mediate HdaB specific adhesive functions.
Keywords: AAF/IV; Escherichia coli; HdaB; adhesion; chaperone-usher; fimbria; invasion; pilus.
© 2016 The Authors Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Figures

Similar articles
-
Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.PLoS Pathog. 2014 Sep 18;10(9):e1004404. doi: 10.1371/journal.ppat.1004404. eCollection 2014 Sep. PLoS Pathog. 2014. PMID: 25232738 Free PMC article.
-
Role of aggregative adherence fimbriae from enteroaggregative Escherichia coli isolates in biofilm and colonization.Microb Pathog. 2025 Jun;203:107444. doi: 10.1016/j.micpath.2025.107444. Epub 2025 Mar 1. Microb Pathog. 2025. PMID: 40032001
-
Crystallization and initial crystallographic analysis of AafA: the major adhesive subunit of the enteroaggregative Escherichia coli AAF/II pilus.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Apr 1;67(Pt 4):454-6. doi: 10.1107/S1744309111001412. Epub 2011 Mar 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21505239 Free PMC article.
-
Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli.Infect Immun. 2015 Apr;83(4):1396-405. doi: 10.1128/IAI.02820-14. Epub 2015 Jan 26. Infect Immun. 2015. PMID: 25624357 Free PMC article.
-
Crystallography and electron microscopy of chaperone/usher pilus systems.Adv Exp Med Biol. 2011;715:159-74. doi: 10.1007/978-94-007-0940-9_10. Adv Exp Med Biol. 2011. PMID: 21557063 Review.
Cited by
-
Structural and functional studies of Escherichia coli aggregative adherence fimbriae (AAF/V) reveal a deficiency in extracellular matrix binding.Biochim Biophys Acta Proteins Proteom. 2017 Mar;1865(3):304-311. doi: 10.1016/j.bbapap.2016.11.017. Epub 2016 Dec 9. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 27939608 Free PMC article.
-
Structural basis of aggregative adherence fimbriae II interactions with sialic acid, mucin, and human intestinal cells.Infect Immun. 2025 Apr 8;93(4):e0048324. doi: 10.1128/iai.00483-24. Epub 2025 Mar 3. Infect Immun. 2025. PMID: 40029240 Free PMC article.
References
-
- Nataro JP, Kaper JB, Robins‐Browne R, Prado V, Vial P, Levine MM (1987) Patterns of adherence of diarrheagenic Escherichia coli to HEp‐2 cells. Pediatr Infect Dis J 6:829–831. - PubMed
-
- Okhuysen PC, Dupont HL (2010) Enteroaggregative Escherichia coli (EAEC): a cause of acute and persistent diarrhea of worldwide importance. J Infect Dis 202:503–505. - PubMed
-
- Nataro JP (2011) Outbreak of hemolytic‐uremic syndrome linked to Shiga toxin‐producing enteroaggregative Escherichia coli O104:H4. Pediatr Res 70:221. - PubMed
-
- Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Muller L, King LA, Rosner B, Buchholz U, Stark K, Krause G, Team HUSI (2011) Epidemic profile of Shiga‐toxin‐producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

