Revisiting overexpression of a heterologous β-glucosidase in Trichoderma reesei: fusion expression of the Neosartorya fischeri Bgl3A to cbh1 enhances the overall as well as individual cellulase activities
- PMID: 27400964
- PMCID: PMC4939661
- DOI: 10.1186/s12934-016-0520-9
Revisiting overexpression of a heterologous β-glucosidase in Trichoderma reesei: fusion expression of the Neosartorya fischeri Bgl3A to cbh1 enhances the overall as well as individual cellulase activities
Abstract
Background: The filamentous fungus Trichoderma reesei has the capacity to secret large amounts of cellulase and is widely used in a variety of industries. However, the T. reesei cellulase is weak in β-glucosidase activity, which results in accumulation of cellobiose inhibiting the endo- and exo-cellulases. By expressing an exogenous β-glucosidase gene, the recombinant T. reesei cellulase is expected to degrade cellulose into glucose more efficiently.
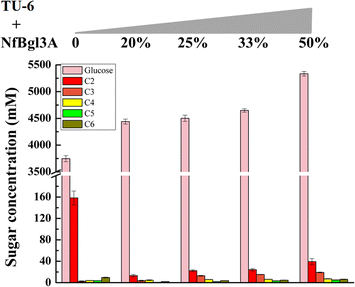
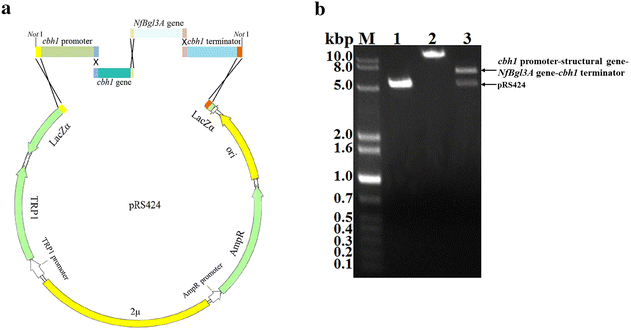
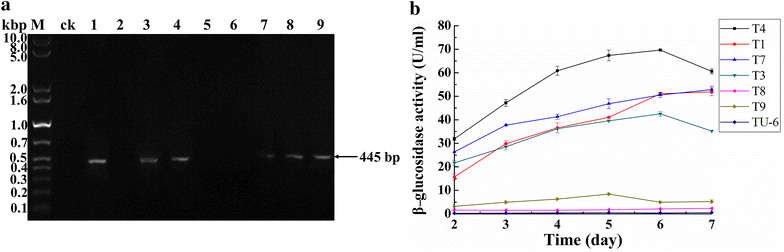
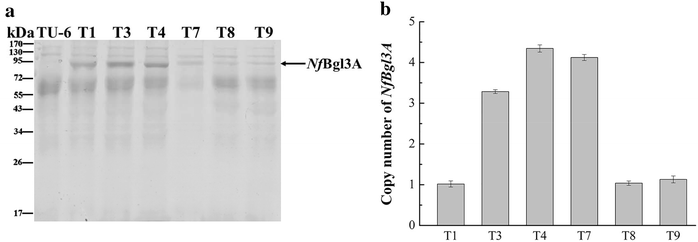
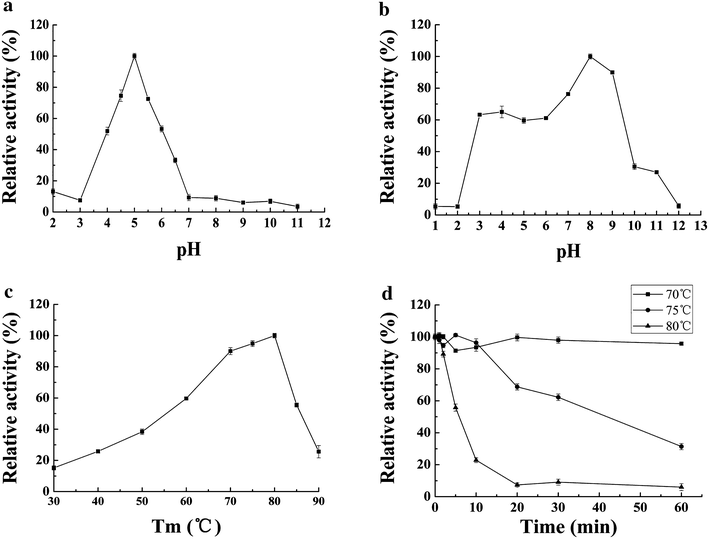
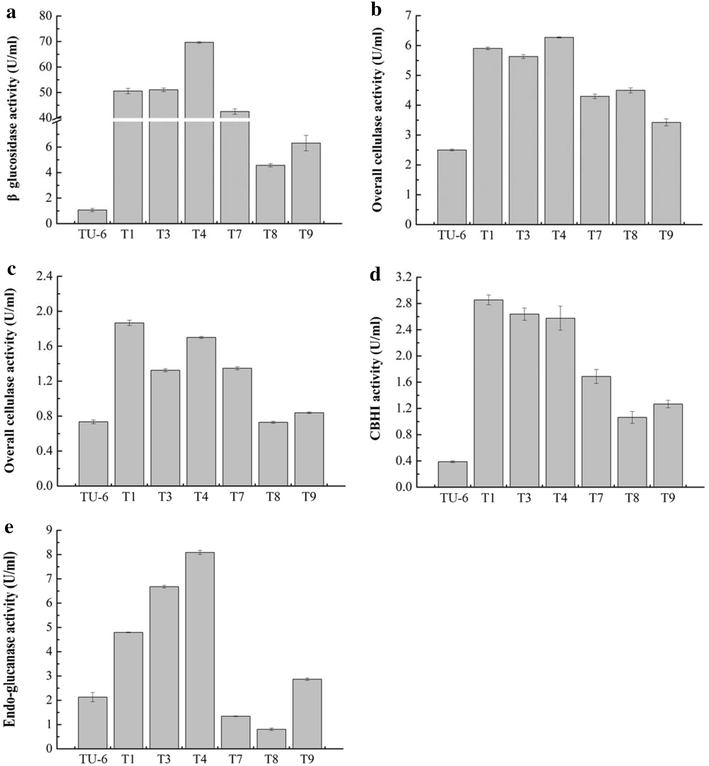
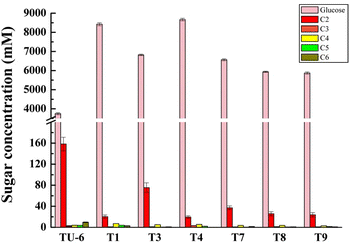
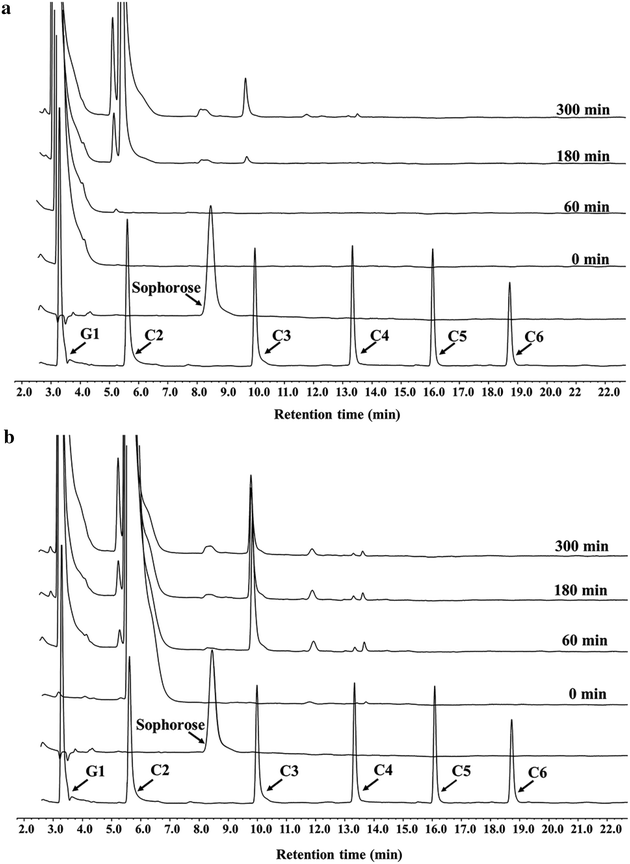
Results: The thermophilic β-glucosidase NfBgl3A from Neosartorya fischeri is chosen for overexpression in T. reesei due to its robust activity. In vitro, the Pichia pastoris-expressed NfBgl3A aided the T. reesei cellulase in releasing much more glucose with significantly lower amounts of cellobiose from crystalline cellulose. The NfBgl3A gene was hence fused to the cbh1 structural gene and assembled between the strong cbh1 promoter and cbh1 terminator to obtain pRS-NfBgl3A by using the DNA assembler method. pRS-NfBgl3A was transformed into the T. reesei uridine auxotroph strain TU-6. Six positive transformants showed β-glucosidase activities of 2.3-69.7 U/mL (up to 175-fold higher than that of wild-type). The largely different β-glucosidase activities in the transformants may be ascribed to the gene copy numbers of NfBgl3A or its integration loci. The T. reesei-expressed NfBgl3A showed highly similar biochemical properties to that expressed in P. pastoris. As expected, overexpression of NfBgl3A enhanced the overall cellulase activity of T. reesei. The CBHI activity in all transformants increased, possibly due to the extra copies of cbh1 gene introduced, while the endoglucanase activity in three transformants also largely increased, which was not observed in any other studies overexpressing a β-glucosidase. NfBgl3A had significant transglycosylation activity, generating sophorose, a potent cellulase inducer, and other oligosaccharides from glucose and cellobiose.
Conclusions: We report herein the successful overexpression of a thermophilic N. fischeri β-glucosidase in T. reesei. In the same time, the fusion of NfBgl3A to the cbh1 gene introduced extra copies of the cellobiohydrolase 1 gene. As a result, we observed improved β-glucosidase and cellobiohydrolase activity as well as the overall cellulase activity. In addition, the endoglucanase activity also increased in some of the transformants. Our results may shed light on design of more robust T. reesei cellulases.
Keywords: Cellulase; DNA assembler; Neosartorya fischeri; Trichoderma reesei; β-Glucosidase.
Figures
References
-
- Sipos B, Benko Z, Dienes D, Reczey K, Viikari L, Siika-aho M. Characterisation of specific activities and hydrolytic properties of cell-wall-degrading enzymes produced by Trichoderma reesei Rut C30 on different carbon sources. Appl Biochem Biotechnol. 2010;161:347–364. doi: 10.1007/s12010-009-8824-4. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

