Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets
- PMID: 27401342
- PMCID: PMC4986526
- DOI: 10.1084/jem.20151467
Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets
Abstract
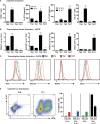
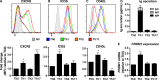
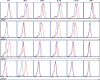
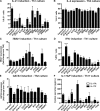
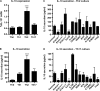
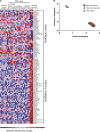
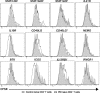
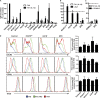
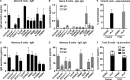
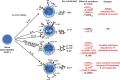
Naive CD4(+) T cells differentiate into specific effector subsets-Th1, Th2, Th17, and T follicular helper (Tfh)-that provide immunity against pathogen infection. The signaling pathways involved in generating these effector cells are partially known. However, the effects of mutations underlying human primary immunodeficiencies on these processes, and how they compromise specific immune responses, remain unresolved. By studying individuals with mutations in key signaling pathways, we identified nonredundant pathways regulating human CD4(+) T cell differentiation in vitro. IL12Rβ1/TYK2 and IFN-γR/STAT1 function in a feed-forward loop to induce Th1 cells, whereas IL-21/IL-21R/STAT3 signaling is required for Th17, Tfh, and IL-10-secreting cells. IL12Rβ1/TYK2 and NEMO are also required for Th17 induction. Strikingly, gain-of-function STAT1 mutations recapitulated the impact of dominant-negative STAT3 mutations on Tfh and Th17 cells, revealing a putative inhibitory effect of hypermorphic STAT1 over STAT3. These findings provide mechanistic insight into the requirements for human T cell effector function, and explain clinical manifestations of these immunodeficient conditions. Furthermore, they identify molecules that could be targeted to modulate CD4(+) T cell effector function in the settings of infection, vaccination, or immune dysregulation.
© 2016 Ma et al.
Figures
References
-
- Avery D.T., Deenick E.K., Ma C.S., Suryani S., Simpson N., Chew G.Y., Chan T.D., Palendira U., Bustamante J., Boisson-Dupuis S., et al. 2010. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207:155–171. 10.1084/jem.20091706 - DOI - PMC - PubMed
-
- Batten M., Ramamoorthi N., Kljavin N.M., Ma C.S., Cox J.H., Dengler H.S., Danilenko D.M., Caplazi P., Wong M., Fulcher D.A., et al. 2010. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. J. Exp. Med. 207:2895–2906. 10.1084/jem.20100064 - DOI - PMC - PubMed
-
- Becattini S., Latorre D., Mele F., Foglierini M., De Gregorio C., Cassotta A., Fernandez B., Kelderman S., Schumacher T.N., Corti D., et al. 2015. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 347:400–406. 10.1126/science.1260668 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

