Tools and techniques for computational reproducibility
- PMID: 27401684
- PMCID: PMC4940747
- DOI: 10.1186/s13742-016-0135-4
Tools and techniques for computational reproducibility
Abstract
When reporting research findings, scientists document the steps they followed so that others can verify and build upon the research. When those steps have been described in sufficient detail that others can retrace the steps and obtain similar results, the research is said to be reproducible. Computers play a vital role in many research disciplines and present both opportunities and challenges for reproducibility. Computers can be programmed to execute analysis tasks, and those programs can be repeated and shared with others. The deterministic nature of most computer programs means that the same analysis tasks, applied to the same data, will often produce the same outputs. However, in practice, computational findings often cannot be reproduced because of complexities in how software is packaged, installed, and executed-and because of limitations associated with how scientists document analysis steps. Many tools and techniques are available to help overcome these challenges; here we describe seven such strategies. With a broad scientific audience in mind, we describe the strengths and limitations of each approach, as well as the circumstances under which each might be applied. No single strategy is sufficient for every scenario; thus we emphasize that it is often useful to combine approaches.
Keywords: Computational reproducibility; Literate programming; Practice of science; Software containers; Software frameworks; Virtualization.
Figures


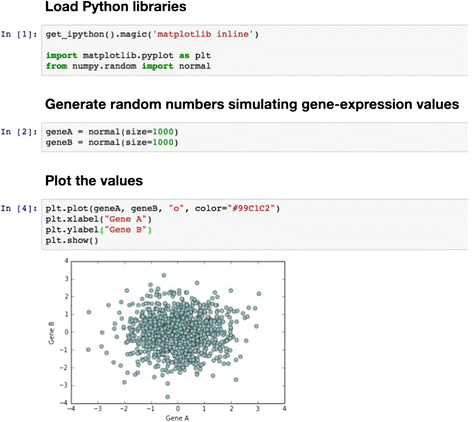

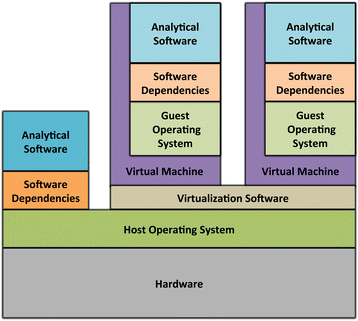
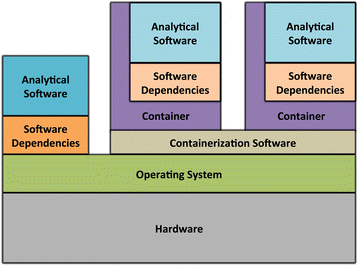

References
-
- Fisher RA. The Design of Experiments. New York: Hafner Press; 1935.
-
- Popper KR. The logic of scientific discovery. London: Routledge; 1959.
-
- Feynman RP. Six Easy Pieces: Essentials of Physics Explained by Its Most Brilliant Teacher. Boston, MA: Addison-Wesley; 1995. p. 34–5.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

