CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome
- PMID: 27402545
- PMCID: PMC4967949
- DOI: 10.15252/embr.201642743
CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome
Abstract
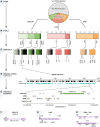
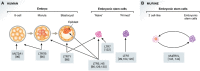
The human genome contains millions of fragments from retrotransposons-highly repetitive DNA sequences that were once able to "copy and paste" themselves to other regions in the genome. However, the majority of retrotransposons have lost this capacity through acquisition of mutations or through endogenous silencing mechanisms. Without this imminent threat of transposition, retrotransposons have the potential to act as a major source of genomic innovation. Indeed, large numbers of retrotransposons have been found to be active in specific contexts: as gene regulatory elements and promoters for protein-coding genes or long noncoding RNAs, among others. In this review, we summarise recent findings about retrotransposons, with implications in gene expression regulation, the expansion of gene isoform diversity and the generation of long noncoding RNAs. We highlight key examples that demonstrate their role in cellular identity and their versatility as markers of cell states, and we discuss how their dysregulation may contribute to the formation of and possibly therapeutic response in human cancers.
Keywords: endogenous retrovirus; lncRNA; regulation; retrotransposon; transcription.
© 2016 The Authors.
Figures

References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

