Internal strain drives spontaneous periodic buckling in collagen and regulates remodeling
- PMID: 27402741
- PMCID: PMC4968713
- DOI: 10.1073/pnas.1523228113
Internal strain drives spontaneous periodic buckling in collagen and regulates remodeling
Abstract
Fibrillar collagen, an essential structural component of the extracellular matrix, is remarkably resistant to proteolysis, requiring specialized matrix metalloproteinases (MMPs) to initiate its remodeling. In the context of native fibrils, remodeling is poorly understood; MMPs have limited access to cleavage sites and are inhibited by tension on the fibril. Here, single-molecule recordings of fluorescently labeled MMPs reveal cleavage-vulnerable binding regions arrayed periodically at ∼1-µm intervals along collagen fibrils. Binding regions remain periodic even as they migrate on the fibril, indicating a collective process of thermally activated and self-healing defect formation. An internal strain relief model involving reversible structural rearrangements quantitatively reproduces the observed spatial patterning and fluctuations of defects and provides a mechanism for tension-dependent stabilization of fibrillar collagen. This work identifies internal-strain-driven defects that may have general and widespread regulatory functions in self-assembled biological filaments.
Keywords: collagenase; matrix metalloproteinase; mechanosensing; pattern formation; single molecule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
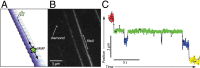
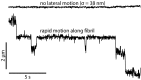
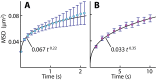




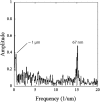
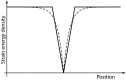
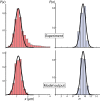
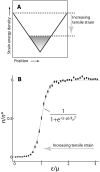
References
-
- Sherman VR, Yang W, Meyers MA. The materials science of collagen. J Mech Behav Biomed Mater. 2015;52:22–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

