Flexibility and Solvation of Amyloid-β Hydrophobic Core
- PMID: 27402826
- PMCID: PMC5000093
- DOI: 10.1074/jbc.M116.740530
Flexibility and Solvation of Amyloid-β Hydrophobic Core
Abstract
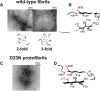
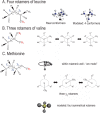
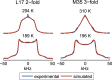
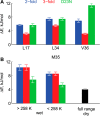
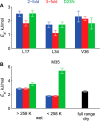
Amyloid fibril deposits found in Alzheimer disease patients are composed of amyloid-β (Aβ) protein forming a number of hydrophobic interfaces that are believed to be mostly rigid. We have investigated the μs-ms time-scale dynamics of the intra-strand hydrophobic core and interfaces of the fibrils composed of Aβ1-40 protein. Using solid-state (2)H NMR line shape experiments performed on selectively deuterated methyl groups, we probed the 3-fold symmetric and 2-fold symmetric polymorphs of native Aβ as well as the protofibrils of D23N Iowa mutant, associated with an early onset of Alzheimer disease. The dynamics of the hydrophobic regions probed at Leu-17, Leu-34, Val-36, and Met-35 side chains were found to be very pronounced at all sites and in all polymorphs of Aβ, with methyl axis motions persisting down to 230-200 K for most of the sites. The dominant mode of motions is the rotameric side chain jumps, with the Met-35 displaying the most complex multi-modal behavior. There are distinct differences in the dynamics among the three protein variants, with the Val-36 site displaying the most variability. Solvation of the fibrils does not affect methyl group motions within the hydrophobic core of individual cross-β subunits but has a clear effect on the motions at the hydrophobic interface between the cross-β subunits, which is defined by Met-35 contacts. In particular, hydration activates transitions between additional rotameric states that are not sampled in the dry protein. Thus, these results support the existence of water-accessible cavity recently predicted by molecular dynamics simulations and suggested by cryo-EM studies.
Keywords: Alzheimer disease; amyloid; amyloid-β (AB); protein dynamic; solid state NMR; solvation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Ahmed A. B., and Kajava A. V. (2013) Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. FEBS Lett. 587, 1089–1095 - PubMed
-
- Cannon M. J., Williams A. D., Wetzel R., and Myszka D. G. (2004) Kinetic analysis of β-amyloid fibril elongation. Anal. Biochem. 328, 67–75 - PubMed
-
- Chimon S., Jones C., Calero D. C., and Ishii Y. (2007) A missing link in amyloid misfolding: structural insights into amyloid intermediates for Alzheimer's β-amyloid, Aβ(1–40) by solid-state NMR. Biophys. J. S, 195A–195A
-
- Fitzpatrick A. W., Debelouchina G. T., Bayro M. J., Clare D. K., Caporini M. A., Bajaj V. S., Jaroniec C. P., Wang L., Ladizhansky V., Müller S. A., MacPhee C. E., Waudby C. A., Mott H. R., De Simone A., Knowles T. P. (2013) Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl. Acad. Sci. U.S.A. 110, 5468–5473 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

