Collagen Accumulation in Osteosarcoma Cells lacking GLT25D1 Collagen Galactosyltransferase
- PMID: 27402836
- PMCID: PMC5000096
- DOI: 10.1074/jbc.M116.723379
Collagen Accumulation in Osteosarcoma Cells lacking GLT25D1 Collagen Galactosyltransferase
Abstract
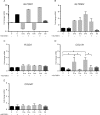
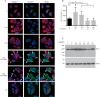
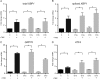
Collagen is post-translationally modified by prolyl and lysyl hydroxylation and subsequently by glycosylation of hydroxylysine. Despite the widespread occurrence of the glycan structure Glc(α1-2)Gal linked to hydroxylysine in animals, the functional significance of collagen glycosylation remains elusive. To address the role of glycosylation in collagen expression, folding, and secretion, we used the CRISPR/Cas9 system to inactivate the collagen galactosyltransferase GLT25D1 and GLT25D2 genes in osteosarcoma cells. Loss of GLT25D1 led to increased expression and intracellular accumulation of collagen type I, whereas loss of GLT25D2 had no effect on collagen secretion. Inactivation of the GLT25D1 gene resulted in a compensatory induction of GLT25D2 expression. Loss of GLT25D1 decreased collagen glycosylation by up to 60% but did not alter collagen folding and thermal stability. Whereas cells harboring individually inactivated GLT25D1 and GLT25D2 genes could be recovered and maintained in culture, cell clones with simultaneously inactive GLT25D1 and GLT25D2 genes could be not grown and studied, suggesting that a complete loss of collagen glycosylation impairs osteosarcoma cell proliferation and viability.
Keywords: CRISPR/Cas; collagen; endoplasmic reticulum (ER); glycosylation; osteosarcoma; protein trafficking (Golgi).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Myllyharju J., and Kivirikko K. I. (2004) Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 20, 33–43 - PubMed
-
- Kukkola L., Hieta R., Kivirikko K. I., and Myllyharju J. (2003) Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J. Biol. Chem. 278, 47685–47693 - PubMed
-
- Vranka J. A., Sakai L. Y., and Bächinger H. P. (2004) Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J. Biol. Chem. 279, 23615–23621 - PubMed
-
- Myllyharju J. (2008) Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann. Med. 40, 402–417 - PubMed
-
- Spiro R. G. (1967) The structure of the disaccharide unit of the renal glomerular basement membrane. J. Biol. Chem. 242, 4813–4823 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

