A Rational Approach for the Identification of Non-Hydroxamate HDAC6-Selective Inhibitors
- PMID: 27404291
- PMCID: PMC4941420
- DOI: 10.1038/srep29086
A Rational Approach for the Identification of Non-Hydroxamate HDAC6-Selective Inhibitors
Abstract
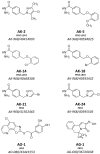
The human histone deacetylase isoform 6 (HDAC6) has been demonstrated to play a major role in cell motility and aggresome formation, being interesting for the treatment of multiple tumour types and neurodegenerative conditions. Currently, most HDAC inhibitors in preclinical or clinical evaluations are non-selective inhibitors, characterised by a hydroxamate zinc-binding group (ZBG) showing off-target effects and mutagenicity. The identification of selective HDAC6 inhibitors with novel chemical properties has not been successful yet, also because of the absence of crystallographic information that makes the rational design of HDAC6 selective inhibitors difficult. Using HDAC inhibitory data retrieved from the ChEMBL database and ligand-based computational strategies, we identified 8 original new non-hydroxamate HDAC6 inhibitors from the SPECS database, with activity in the low μM range. The most potent and selective compound, bearing a hydrazide ZBG, was shown to increase tubulin acetylation in human cells. No effects on histone H4 acetylation were observed. To the best of our knowledge, this is the first report of an HDAC6 selective inhibitor bearing a hydrazide ZBG. Its capability to passively cross the blood-brain barrier (BBB), as observed through PAMPA assays, and its low cytotoxicity in vitro, suggested its potential for drug development.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

