One size fits all? Direct evidence for the heterogeneity of genetic drift throughout the genome
- PMID: 27405384
- PMCID: PMC4971176
- DOI: 10.1098/rsbl.2016.0426
One size fits all? Direct evidence for the heterogeneity of genetic drift throughout the genome
Abstract
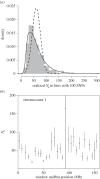
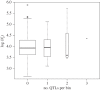
Effective population size (Ne) is a central parameter in population and conservation genetics. It measures the magnitude of genetic drift, rates of accumulation of inbreeding in a population, and it conditions the efficacy of selection. It is often assumed that a single Ne can account for the evolution of genomes. However, recent work provides indirect evidence for heterogeneity in Ne throughout the genome. We study this by examining genome-wide diversity in the Danish Holstein cattle breed. Using the differences in allele frequencies over a single generation, we directly estimated Ne among autosomes and smaller windows within autosomes. We found statistically significant variation in Ne at both scales. However, no correlation was found between the detected regional variability in Ne, and proxies for the intensity of linked selection (local recombination rate, gene density), or the presence of either past strong selection or current artificial selection on traits of economic value. Our findings call for further caution regarding the wide applicability of the Ne concept for understanding quantitatively processes such as genetic drift and accumulation of consanguinity in both natural and managed populations.
Keywords: Holstein breed; effective population size; genetic drift; linked selection; quantitative trait loci.
© 2016 The Author(s).
Figures
Similar articles
-
Trends in genome-wide and region-specific genetic diversity in the Dutch-Flemish Holstein-Friesian breeding program from 1986 to 2015.Genet Sel Evol. 2018 Apr 11;50(1):15. doi: 10.1186/s12711-018-0385-y. Genet Sel Evol. 2018. PMID: 29642838 Free PMC article.
-
Genome-wide genetic diversity of Holstein Friesian cattle reveals new insights into Australian and global population variability, including impact of selection.Anim Genet. 2007 Feb;38(1):7-14. doi: 10.1111/j.1365-2052.2006.01543.x. Anim Genet. 2007. PMID: 17257182
-
What Is Ne, Anyway?J Hered. 2022 Jul 23;113(4):371-379. doi: 10.1093/jhered/esac023. J Hered. 2022. PMID: 35532202
-
Prediction and estimation of effective population size.Heredity (Edinb). 2016 Oct;117(4):193-206. doi: 10.1038/hdy.2016.43. Epub 2016 Jun 29. Heredity (Edinb). 2016. PMID: 27353047 Free PMC article. Review.
-
Genetic drift, selection and the evolution of the mutation rate.Nat Rev Genet. 2016 Oct 14;17(11):704-714. doi: 10.1038/nrg.2016.104. Nat Rev Genet. 2016. PMID: 27739533 Review.
Cited by
-
Heterogeneity in effective size across the genome: effects on the inverse instantaneous coalescence rate (IICR) and implications for demographic inference under linked selection.Genetics. 2022 Mar 3;220(3):iyac008. doi: 10.1093/genetics/iyac008. Genetics. 2022. PMID: 35100421 Free PMC article.
-
Fishing for DNA? Designing baits for population genetics in target enrichment experiments: Guidelines, considerations and the new tool supeRbaits.Mol Ecol Resour. 2022 Jul;22(5):2105-2119. doi: 10.1111/1755-0998.13598. Epub 2022 Mar 3. Mol Ecol Resour. 2022. PMID: 35178874 Free PMC article.
-
Using singleton densities to detect recent selection in Bos taurus.Evol Lett. 2021 Nov 22;5(6):595-606. doi: 10.1002/evl3.263. eCollection 2021 Dec. Evol Lett. 2021. PMID: 34917399 Free PMC article.
-
Whole-genome sequencing of cryopreserved resources from French Large White pigs at two distinct sampling times reveals strong signatures of convergent and divergent selection between the dam and sire lines.Genet Sel Evol. 2023 Mar 2;55(1):13. doi: 10.1186/s12711-023-00789-z. Genet Sel Evol. 2023. PMID: 36864379 Free PMC article.
-
The estimates of effective population size based on linkage disequilibrium are virtually unaffected by natural selection.PLoS Genet. 2022 Jan 25;18(1):e1009764. doi: 10.1371/journal.pgen.1009764. eCollection 2022 Jan. PLoS Genet. 2022. PMID: 35077457 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

